Abstract
Arisaema bockii Engler is a perennial herbaceous medicinal plant, which is widely distributed in many provinces in China such as Anhui, Jiangsu, and Zhejiang. In this study, the complete chloroplast genome sequence of A. bockii was assembled and characterized based on high-throughput sequencing data. The total length of chloroplast genome was 175,537 bp, including large single-copy (LSC) and small single-copy (SSC) regions of 98,870 bp and 23,345 bp, respectively, which were separated by a pair of 27,161 bp inverted repeat (IR) regions. The genome contained 129 genes, including 84 protein-coding genes, 36 tRNA genes, 8 rRNA genes, and one pseudogene. The overall GC content of the genome was 33.6%. A phylogenetic tree reconstructed by 30 chloroplast genomes revealed that A. bockii was mostly related to the same genus species A. ringens, A. franchetianum and A. erubescens. The work reported the first complete chloroplast genome of A. bockii, which may provide some useful information to the evolution of the family Araceae.
Arisaema bockii Engler is a kind of Chinese herbal medicine, which is widely distributed in China and south of Japan. It is one of the most beautiful species in the Arisaema genus of the family Araceae (Hayakawa et al. Citation2011). Its medicinal parts are tubers, which can be used as the substitute for A. erubescens, A. heterophyllum, and A. amurense listed in Chinese Pharmacopeia with the function of dissipating binds and dispersing swelling (Wang et al. Citation2012). Pharmacological analysis indicated that A. bockii has the activities of anti-tumor (Feng et al. Citation2016), anti-bacterial (Wang et al. Citation2012), and nematicidal (Du et al. Citation2011). Flavonoids, terpenoids, alkaloids and glycosphingolipids are its main medicinal ingredients (Kant et al. Citation2020). However, little is known about its biosynthesis mechanism of the compounds. In addition, its phylogenetic position is not very clear causing on the lack of genomic information. Here, we characterized the complete chloroplast genome sequence of A. bockii using high throughput sequencing technology, which will provide useful informatics data for the phylogeny of A. bockii and other related species.
The fresh leaves of A. bockii were collected from Lu’an, Anhui, China (31°77′ N, 115°93′ E). Specimens were stored in the Herbarium of West Anhui University (the accession number is WAU-DTL-20210218-1, and the email of the sample contact is [email protected]). Total genomic DNA was extracted from the leaves according to a modified CTAB protocol (Doyle and Doyle Citation1987). The DNA was stored at −80 °C in our lab. The whole genome sequencing was conducted by Hefei Biodata Biotechnologies Inc. (Hefei, China) on the Illumina Hiseq platform. The filtered sequences were assembled by using the program SPAdes assembler 3.10.0 (Bankevich et al. Citation2012). The chloroplast genes were annotated by GeSeq (Tillich et al. Citation2017) with default parameters to predict protein-coding genes, rRNA and tRNA genes. All tRNA genes were further verified by using tRNAscan-SE 2.0 (Chan et al. Citation2021).
The chloroplast genome of A. bockii was determined to comprise a 175,537 bp, double stranded, and circular DNA (GenBank accession no. MZ380241), containing two inverted repeat (IR) regions of 27,161 bp, and separated by large single-copy (LSC) and small single-copy (SSC) regions of 98,870 bp and 23,345 bp, respectively. The genome was predicted to have 129 genes, including 84 protein-coding genes, 36 tRNA genes, 8 rRNA genes, and one pseudogene. Five protein-coding genes, seven tRNA genes and four rRNA genes were duplicated in IR regions. Nineteen genes contained two exons and four genes (clpP, ycf3 and two rps12) contained three exons. The overall GC content of A. bockii cp genome was 33.6% and the corresponding values in LSC, SSC and IR regions were 31.6%, 27.4% and 39.9%, respectively.
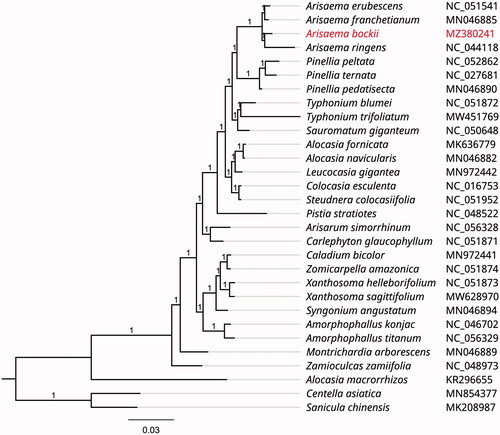
To investigate its taxonomic status, alignment was performed with 30 reported chloroplast genome sequences of the family Araceae (Centella asiatica and Sanicula chinensis were used as outgroup) using MAFFT v7.307 (Katoh and Standley Citation2013), and then a maximum likelihood (ML) tree was constructed by FastTree version 2.1.10 with Generalized Time-Reversible (GTR) model, 1000 bootstrap replicates (Price Citation2010). As expected, A. bockii was mostly related to the same genus species A. ringens, A. franchetianum and A. erubescens with bootstrap support values of 100% ().
Figure 1. Phylogenetic tree inferred by Maximum Likelihood (ML) method based on 30 representative species. Centella asiatica and Sanicula chinensis were used as an outgroup. A total of 1000 bootstrap replicates were computed and the bootstrap support values were shown at the branches. GenBank accession numbers were shown in .
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data of A. bockii that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ380241. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA737041, SRR14793494, and SAMN19678498, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Chan PP, Lin BY, Mak AJ, Lowe TM. 2021. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 49:9077–9096.
- Du SS, Zhang HM, Bai CQ, Wang CF, Liu QZ, Liu ZL, Wang YY, Deng ZW. 2011. Nematocidal flavone-C-glycosides against the root-knot nematode (Meloidogyne incognita) from Arisaema erubescens tubers. Molecules. 16(6):5079–5086.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Feng LX, Sun P, Mi T, Liu M, Liu W, Yao S, Cao YM, Yu XL, Wu WY, Jiang BH, Yang M, et al. 2016. Agglutinin isolated from Arisema heterophyllum Blume induces apoptosis and autophagy in A549 cells through inhibiting PI3K/Akt pathway and inducing ER stress. Chin J Nat Med. 14(11):856–864.
- Hayakawa H, Hamachi H, Matsuyama K, Muramatsu Y, Minamiya Y, Ito K, Yokoyama J, Fukuda T. 2011. Interspecific hybridization between Arisaema sikokianum and A. serratum (Araceae) confirmed through nuclear and chloroplast DNA comparisons. AJPS. 02(04):521–526.
- Kant K, Lal UR, Rawat R, Kumar A, Ghosh M. 2020. Genus Arisaema: a review of traditional importance, chemistry and biological activities. Comb Chem High Throughput Screen. 23(7):624–648.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2-approximately maximum-likelihood trees for large alignments. PloS One. 5(3):e9490.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang LW, Xu BG, Wang JY, Su ZZ, Lin FC, Zhang CL, Kubicek CP. 2012. Bioactive metabolites from Phoma species, an endophytic fungus from the Chinese medicinal plant Arisaema erubescens. Appl Microbiol Biotechnol. 93(3):1231–1239.
