Abstract
Phlomoides rotata (Benth. ex Hook.f.) Mathiesen is a perennial herb endemic to Qinghai-Tibet Plateau with important medicinal properties. Here, we sequenced and analyzed the complete chloroplast (cp) genome of P. rotata and reconstructed the phylogeny of P. rotata based on 24 cp genomes. The genome of P. rotata is 151,825 bp in length, including a large single-copy (LSC) region of 83,129 bp and a small single-copy (SSC) region of 17,398 bp. A total of 131 genes were identified, of which 86 are protein-coding genes, 37 are transfer RNA genes, and eight are ribosomal RNA genes. Phylogenetic analyses revealed that the species P. rotata is closely related to Phlomoides alpina with bootstrap support (BS) values of 100%. Overall, the genomic resources presented in this study will be beneficial for further studies on evolutionary patterns of P. rotata and its closely related species.
Phlomoides rotata (Benth. ex Hook.f) Mathiesen (P. rotata), a perennial herb belonging to the family Lamiaceae, is endemic to the Qinghai-Tibet Plateau and widely distributes in 2700–4900 m weathered alpine alluvial fans, stony alpine meadows, and floodplains (Wu and Li Citation1977). As a traditional Tibetan medicinal herb, it is used to treat the traumatic injury, promote blood circulation, and alleviate pain (Liu et al. Citation2006, Li et al. Citation2008). However, its classification is currently highly controversial. In this study, the complete chloroplast (cp) genome of this species was sequenced and annotated, which might be useful as a genetic resource for plant identification and further phylogenetic study.
Total genomic DNA was extracted from P. rotata leaves collected from Lhasa, Tibet, China (N29°46’41ʺ, E91°10’19ʺ) using the modified CTAB method (Murray and Thompson Citation1980). The voucher specimen (2020MaC24) was deposited in the Herbarium of Tibet Plateau Institute of Biology (Chao Ma, [email protected]). Paired-end (150 bp) sequencing was conducted in Illumina HiSeq 2500 platform, and the cp reads were filtered using NGS-QC toolkit (Patel and Jain, Citation2012) and were assembled by using the program SPAdes 3.11.0 (Bankevich et al. Citation2012) with a k-mer set of 93, 105, 117, and 121. Genome annotation was performed by Plann v1.1.2 (Huang and Cronk Citation2015), Aragorn v1.2.38 (Laslett and Canback Citation2004) and Blast v2.6.0. Phlomoides betonicoides (MN617020.1) cp genome sequence was used as the reference sequence.
The complete cp genome of P. rotata (MZ150795.1) is a typical quadripartite structure of 151,825 bp, consisting of a large single-copy (LSC) region of 83,129 bp, a small single-copy (SSC) region of 17,398 bp, and a pair of inverted repeats (IR) region of 25,649 bp. The assembled cp genome has 131 annotated genes, including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Among those, 18 are duplicated in IR regions, including seven protein-coding genes, seven tRNA genes, and four rRNA genes. In addition, there are 15 genes with single intron (trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC, ndhA) and 2 genes (clpP and ycf3) with two introns. The base composition of the complete cp genome sequence was analyzed and found to be 38.46% GC content. Furthermore, the GC content of IR regions was 43.40%, which is higher than that of the LSC (36.65%)/SSC (32.53%).
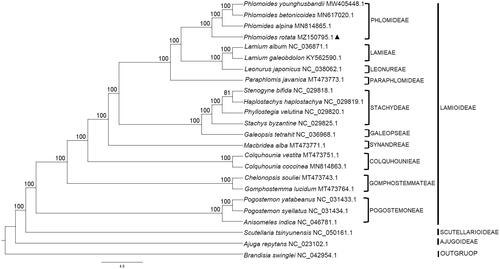
To investigate its phylogenetic placement in the family, a total of 23 complete cp genomes including one outgroup (Brandisia swinglei) were downloaded from NCBI GenBank and aligned with the cp genome of P. rotata by using MAFFT (Katoh and Stanley Citation2013). The maximum likelihood (ML) analysis using GTR-I-G model was performed in RaxML v8.1.11 (Stamatakis, Citation2014). ML consensus tree revealed that P. rotata occupies the basal position in the Phlomoides clade (BS = 100%, ). It diverged earlier than the rest of the Phlomoides but it has a higher affinity to Phlomoides alpina. The complete cp genome sequence of P. rotata is expected to provide new insights into the evolutionary history of Lamiaceae and its siblings.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/nuccore/MZ150795.1) under the accession no. MZ150795.1. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA727957, SAMN19066089 and SRR14470730, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Huang DI, Cronk QC. 2015. Plann: A command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 30: 772–780.
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16.
- Li M, Jia Z, Hu Z, Zhang R, Tao S. 2008. Experimental study on the hemostatic activity of the Tibetan medicinal herb Lamiophlomis rotata. Phytother Res. 22(6):759–765.
- Liu J, Li W, Geng Y, Wang Q, Luo L, Yang Z. 2006. Genetic diversity and population structure of Lamiophlomis rotata (Lamiaceae), an endemic species of Qinghai-Tibet Plateau. Genetica. 128(1–3):385–394.
- Murray MG, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucl Acids Res. 8(19):4321–4326.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7(2):e30619.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wu CY, Li XW. 1977. Flora of China. Vol. 65(2). Beijing: Science Press.