Abstract
Calohypnum plumiforme is a widely distributed moss in East Asia. In this study, Illumina sequencing data was used to assemble the complete chloroplast genome of C. plumiforme. The length of the circular genome is 124,037 bp. It contains a total of 121 genes, including 81 protein-coding, 36 tRNA, and 4 rRNA genes. The GC content of the chloroplast genome of C. plumiforme is 29.07%. The phylogenetic analysis suggested that C. plumiforme is sister to Calliergonella cuspidata. This study provides important sequence information for species identification and its phylogenetic relationship in the Bryopsida.
Calohypnum plumiforme is a relatively large moss which is frequently used for landscape construction and as an indicator of environmental pollution (Oguri & Deguchi et al. Citation2018; Wierzcholska et al. Citation2018; Zhang et al. Citation2020). It belongs to the family Hypnaceae which are difficult to classify. As a representative species which are widely distributed in East Asia, C. plumiforme occurs in various habitats and on different substrates, due to its wide ecological range. The complete chloroplast genome sequence of C. plumiforme has not been published so far. Herein, we report high-throughput sequencing of a C. plumiforme specimen to determine its chloroplast (cp) genome structure and evolutionary relationships within the Bryopsida.
The fresh leaves of C. plumiforme were collected from Lishui Runsheng Moss Technology Co. Ltd, in Zhejiang Province, China (28°29′27″N, 119°54′34″E). The voucher specimen of C. plumiforme was deposited at the Lishui University Herbarium (http://www.lsu.edu.cn/) under the voucher number LS-SAM2020-11A. The genomic DNA was extracted using Plant Genomic DNA Kit, DP305 (TIANGEN, Beijing, China). The sequencing library was produced using the Illumina Truseq™ DNA Sample Preparation Kit (Illumina, San Diego, USA) according to the manufacturer's recommendations. The prepared library was loaded on the Illumina Novaseq 6000 platform for PE 2 × 150 bp sequencing at Novogene (Beijing, China). The raw data were used to assemble the complete cp genome using GetOrganelle (Jin et al. Citation2020). Genome annotation was performed with PGA (Qu et al. Citation2019) by comparing the sequences with the cp genome of Amblystegium serpens, GeneBank Accession Number NC_049069 (Wei et al. Citation2020). The annotated genome sequence was deposited in GenBank under Accession Number MZ297475.
The circular cp genome of C. plumiforme was 124,037 bp in length, containing a larger single copy (LSC) region of 86,011 bp in length, a smaller single copy (SSC) region of 18,530 bp and two inverted repeats (IRs), each 9,748 bp. The total GC content is 29.07%, while the GC content of the LSC, SSC and IRs regions are 26.30%, 25.72% and 44.5%, respectively. A total of 121 unique genes were predicted, including 81 protein-coding, 36 tRNA and 4 rRNA genes.
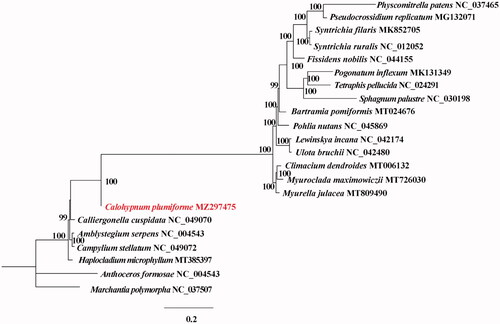
A phylogenetic analysis was performed using complete cp genomes from 20 Bryopsida species with Marchantia polymorpha and Anthoceros formosae serving as the outgroup Taxa. The genomes were aligned with the MAFFT v7.388 using default settings (Katoh and Standley Citation2013). The phylogenetic analysis was conducted based on maximum likelihood (ML) analyses implemented in IQ-TREE v2.1.2 with the TVM + F+R5 nucleotide substitution model, which was selected by ModelFinder (Kalyaanamoorthy et al. Citation2017; Minh et al. Citation2020). The support for the inferred ML tree was inferred by bootstrapping with 1,000 replicates. The phylogenetic analysis suggested that C. plumiforme is sister to Calliergonella cuspidata (). This study provides important sequence information for species identification, and its phylogenetic relationship in Bryopsida.
Figure 1. Maximum-likelihood (ML) tree based on 20 cp genome sequences of representative Bryopsida and Anthoceros formosae and Marchantia polymorpha were designated as outgroup. Numbers on the nodes are bootstrap values based on 1,000 replicates. The C. plumiforme genome was marked in bold and red font.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no MZ297475. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA732638, SAMN19328312 and SRR14642712, respectively.
Additional information
Funding
References
- Jin JJ, Yu WB, Yang JB, Song Y, Depamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1–31.
- Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Oguri E, Deguchi H. 2018. Radiocesium contamination of the moss Hypnum plumaeforme caused by the Fukushima Dai-Ichi nuclear power plant accident. J Environ Radioact. 192:648–653.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Wei S, Yue XR, Li N, Liu Y, Wu YH. 2020. Comparison of eight complete plastid genomes from three moss families Amblystegiaceae, Calliergonaceae and Pylaisiaceae. Mitochondrial DNA Part B. 5(3):3109–3111.
- Wierzcholska S, Dyderski MK, Pielech R, Gazda A, Smoczyk M, Malicki M, Horodecki P, Kamczyc J, Skorupski M, Hachułka M, et al. 2018. Natural forest remnants as refugia for bryophyte diversity in a transformed mountain river valley landscape. Sci Total Environ. 640–641:954–964.
- Zhang Y, Li C, Luo X. 2020. Enrichment effect of Hypnum plumaeforme on 210Po and 210Pb. Int J Phytoremediation. 22(2):140–147.
