Abstract
The plastid genome of the deep-shade plant Selaginella erythropus, which has highly unusual chloroplasts, was characterized using Illumina pair-end sequencing. This plastome is 140,151 bp in length with a large single-copy region (LSC) of 56,133 bp, a small single-copy region (SSC) of 61,268 bp, and two direct repeats (DRs) of 11,375 bp. The overall GC content is 50.68%, while those of LSC, SSC, and DR are 48.96%, 50.3%, and 55.96%, respectively. The plastome contains 102 genes, including 76 protein-coding, 15 tRNA (12 tRNA species), and 8 rRNA genes (4 rRNA species). The phylogenetic analysis shows that S. erythropus is closely related to S. moellendorffii and S. doederleinii. This result is consistent with the previous phylogenetic relationship inferred from multiple plastid and nuclear loci. However, only S. erythropus has the two-zoned giant chloroplast, the bizonoplast. The plastome provides an excellent reference for understanding the unique chloroplast differentiation in Selaginellaceae.
In most land plants, chloroplasts are located in the mesophyll cells, with 50‒250 chloroplasts per cell (Pyke Citation2009). Selaginella is unusual in having several variations on this typical chloroplast structure (Liu et al. Citation2020) including giant chloroplasts of several different forms. The bizonoplast, found in every leaf dorsal epidermal cell of some Selaginella species, is a cup-shaped giant chloroplast with dimorphic ultrastructure (Sheue et al. Citation2007). The upper zone is characterized by groups of 2‒4 thylakoid membranes parallel to each other, while the lower zone consists of grana and stroma thylakoid membranes, similar to normal chloroplasts.
The bizonoplast, first reported from S. erythropus (Mart.) Spring (Sheue et al. Citation2007), and only reported in Selaginella, originates from a proplastid, developing its zoned structure after exposure to low light (Sheue et al. Citation2015). Selaginella, with about 750 species occurring globally in various habitats (Jermy Citation1990), is an ideal model genus to understand the diversity of chloroplasts and their adaptive significance. Given the unique structure of the bizonoplast and its environmental correlates, the chloroplast genome of a bizonoplast-containing species is of special interest. Here we assembled and annotated the plastid genome of S. erythropus from a specimen growing in the nursery at National Chung Hsing University, Taiwan (24° 07′N, 120° 40′E) to contribute to the bioinformatics and genome structure of the bizonoplast.
The total genomic DNA was extracted from fresh shoots of S. erythropus (voucher # Liu JW-05, in the herbarium of National Chung Hsing University, TCB, Taiwan; Chiou-Rong Sheue, [email protected]) using the CTAB method (Doyle and Doyle Citation1990). The genomic DNA was fragmented and libraries were constructed with the insertion sizes 180 bp, 350 bp, 500 bp, and 700 bp. These libraries were paired-end sequenced on an Illumina HiSeq platform (Illumina Inc., San Diego, CA). The de novo assembly was performed using the GENEIOUS Prime Velvet plugin (Zerbino and Birney Citation2008), and subsequently, the plastid contigs were arranged based on the plastome of S. moellendorffii (HM173080) (Banks et al. Citation2011). PCR and Sanger sequencing were conducted to confirm the sequences from the SC-DR junctions and highly variable regions. The S. erythropus plastome was annotated using the software PGA (Qu et al. Citation2019) and GENEIOUS Prime (Kearse et al. Citation2012) by comparing it with the plastomes of S. moellendorffii (HM173080) (Banks et al. Citation2011) and S. doederleinii (MH598532) (Zhang et al. Citation2019).
Typically, plastomes contain two inverted repeats (IRs) separated by a large single-copy region (LSC) and a small single-copy region (SSC) (Mower and Vickrey Citation2018). However, the S. erythropus plastome features a set of direct repeats (DRs), similar to those from some other Selaginella species (Mower et al. Citation2019; Zhang et al. Citation2019) (). The plastome is 140,151 bp in length and has two DRs of 11,375 bp (8.12%), which are separated by an LSC of 56,133 bp (40.05%) and an SSC of 61,268 bp (43.72%). The DR in Selaginella species is presumably caused by a large inversion containing a former IR copy (i.e. IRb). This inversion also relocated a partial LSC region into the former SSC region, thereby resulting in the relatively short LSC as compared to the SSC in other Selaginella (Shim et al. Citation2021).
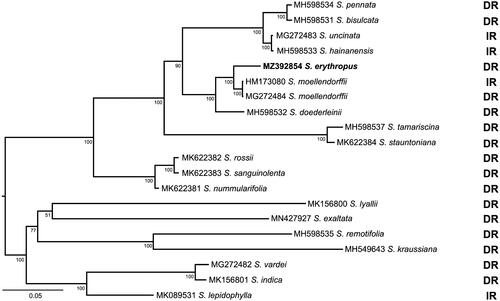
Figure 1. The maximum likelihood (ML) tree of 20 sampled Selaginella species. Bootstrap proportion (BP) values are indicated along branches. The bold taxon is S. erythropus, which is newly sequenced in this study. The scale bar denotes 0.05 substitutions per nucleotide. DR: direct repeats; IR: inverted repeats.
The S. erythropus plastome GC content is 50.68%, with the LSC, SSC, and DR regions GC contents being 48.96%, 50.3%, and 55.96%, respectively. The plastome GC content of S. erythropus is on the low end of the range (50.75%−56.49%) from 16 other Selaginella species (Shim et al. Citation2021) but is much higher than the average for 3,507 plastome sequences from algae to seed plants (37.38 ± 2.26%, Kwon et al. Citation2020). The S. erythropus plastome comprises 102 genes, including 76 protein-coding (76 PCG species), 8 ribosomal RNA (4 rRNA species) and 15 transfer RNA genes (12 tRNA species). Nine PCG genes (atpF, clpP, ndhA, ndhB, petB, petD, rpl2, rpl16 and rpoC1) harbor a single intron, and one (ycf3) contains two introns. In addition, accD, infA and rpl20 are likely pseudogenes because of incomplete open reading frames.
To construct a phylogenetic tree, 51 shared protein-coding genes of 20 plastomes were extracted and aligned individually in MAFFT (Katoh and Standley Citation2013) implemented in GENEIOUS Prime (Kearse et al. Citation2012). The maximum likelihood (ML) tree was determined using GARLI v.2.0 (Zwickl Citation2006), with 1000 bootstrap replicates and the best model GTR + G + I model was selected based on Akaike Information Criterion (AIC) in jModeltest (Posada Citation2008). In the ML tree, S. erythropus is most closely related to S. moellendorffii (). This result is consistent with the previous phylogenetic relationship inferred from three gene regions (rbcL, pgiC, SQD1) and morphological features (Weststrand and Korall Citation2016). The plastome of S. erythropus provides an excellent reference for elucidating the evolution and functional divergence of the giant chloroplasts and bizonoplasts in Selaginellaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ392854. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA752173, SUB10127488, and SAMN20585740, respectively.
Additional information
Funding
References
- Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M, DePamphilis C, Albert VA, Aono N, Aoyama T, Ambrose BA, et al. 2011. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 332(6032):960–963.
- Doyle JJ, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus. 12(13):39–40.
- Jermy AC. 1990. Selaginellaceae. In: Kramer KU, Green PS, editors. The families and genera of vascular plants, vol. I. Pteridophytes and gymnosperms. Berlin, Germany: Springer-Verlag; p. 39–45.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kwon E-C, Kim J-H, Kim N-S. 2020. Comprehensive genomic analyses with 115 plastomes from algae to seed plants: structure, gene contents, GC contents, and introns. Genes Genomics. 42(5):553–570.
- Liu J-W, Li S-F, Wu C-T, Valdespino IA, Ho J-F, Wu Y-H, Chang H-M, Guu T-Y, Kao M-F, Chesson C, et al. 2020. Gigantic chloroplasts, including bizonoplasts, are common in shade-adapted species of the ancient vascular plant family Selaginellaceae. Am J Bot. 107(4):562–576.
- Mower JP, Ma P-F, Grewe F, Taylor A, Michael TP, VanBuren R, Qiu Y-L. 2019. Lycophyte plastid genomics: extreme variation in GC, gene and intron content and multiple inversions between a direct and inverted orientation of the rRNA repeat. New Phytol. 222(2):1061–1075.
- Mower JP, Vickrey TL. 2018. Structural diversity among plastid genomes of land plants. In: Chaw S-M, Jansen RK, editors. Advances in botanical research. Vol. 85. Cambridge, MA, USA: Academic Press; p. 263–292.
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25(7):1253–1256.
- Pyke K. 2009. Plastid biology. Cambridge, UK: Cambridge University Press.
- Qu X-J, Moore MJ, Li D-Z, Yi T-S. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Sheue C-R, Liu J-W, Ho J-F, Yao A-W, Wu Y-H, Das S, Tsai C-C, Chu H-A, Ku MSB, Chesson P. 2015. A variation on chloroplast development: the bizonoplast and photosynthetic efficiency in the deep-shade plant Selaginella erythropus. Am J Bot. 102(4):500–511.
- Sheue C-R, Sarafis V, Kiew R, Liu H-Y, Salino A, Kuo‐Huang L-L, Yang Y-P, Tsai C-C, Lin C-H, Yong JWH, et al. 2007. Bizonoplast, a unique chloroplast in the epidermal cells of microphylls in the shade plant Selaginella erythropus (Selaginellaceae). Am J Bot. 94(12):1922–1929.
- Shim H, Lee HJ, Lee J, Lee HO, Kim J-H, Yang T-J, Kim N-S. 2021. Plastid genomes of the early vascular plant genus Selaginella have unusual direct repeat structures and drastically reduced gene numbers. IJMS. 22(2):641.
- Weststrand S, Korall P. 2016. Phylogeny of Selaginellaceae: there is value in morphology after all!. Am J Bot. 103(12):2136–2159.
- Zerbino BR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhang H-R, Xiang Q-P, Zhang X-C. 2019. The unique evolutionary trajectory and dynamic conformations of DR and IR/DR-coexisting plastomes of the early vascular plant Selaginellaceae (Lycophyte). Genome Biol Evol. 11(4):1258–1274.
- Zwickl DJ. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion [Ph.D. dissertation]. The University of Texas at Austin.
