Abstract
In the present study, we announce the first complete chloroplast genome sequence of Artocarpus tonkinensis, a tree native to China with diverse beneficial uses. This complete chloroplast genome is 160,987 bp in length. In total, 130 genes were identified, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The findings of phylogenetic analysis supported that Artocarpus belongs to the Moraceae family and proposed a sister relationship between Artocarpus and Morus.
Artocarpus tonkinensis A. Chev. ex Gagnep. belongs to the genus Artocarpus J. R. Forst. & G. Forst. (Family: Moraceae) (Zhou and Michael Citation2003)). It is an evergreen tree that is up to 14–16 m high and is mainly distributed in various countries such as Cambodia, northern Vietnam, and China (Fujian, Guangdong, Guangxi, Guizhou, Hainan, and southern Yunnan) (Zhang et al. Citation1998; Zhou and Michael Citation2003). A. tonkinensis has very hardwood, and its fruits are edible and sweet in taste (Zhang et al. Citation1998; Zhou and Michael Citation2003). It has a variety of beneficial medicinal uses; its leaf decoction is used as a treatment for joint disorders and backache in northern Vietnam (Thuy et al. Citation2017; Orecchini et al. Citation2020). Besides, it is also an ornamental street tree with a big and dense canopy. In the present study, we assembled and characterized the complete chloroplast genome of A. tonkinensis. These findings will enrich the gene information and certainly contribute to the further study of this species.
The fresh leaves of A. tonkinensis were collected from Qinzhou City (Guangxi, China; 22° 17’46″N, 108°41′47″E). Voucher specimens were deposited at the herbarium of Guangxi Forestry Research Institute (Mr. Li, [email protected]) (registration number: 2021081302), whereas the DNA samples were stored at Guangxi Key Laboratory of Special Non-wood Forest Cultivation and Utilization (Nanning, China). A sample’s total genomic DNA was extracted from about 100 mg fresh leaves by using a modified CTAB method (Doyle and Doyle Citation1987). Constructed the libraries with an average length of 350 bp by using the NexteraXT DNA Library Preparation Kit (Illumina, San Diego, CA), then the libraries were sequenced on Illumina Novaseq 6000 platform. Raw sequence reads were edited by using NGS QC Tool kit (Patel and Jain Citation2012), clean data were de novo assembled by SPAdes v.3.11.0 software (Bankevich et al. Citation2012). Finally, the assembled complete chloroplast genome was annotated via PGA (Qu et al. Citation2019) and submitted to GenBank (accession number: MZ379793).
The length of the complete chloroplast genome of A. tonkinensis was 160,987 bp, with a total GC content of 35.8%. The complete chloroplast consisted of a large single copy (LSC) region of 89,551 bp, a small single copy (SSC) region of 20,072 bp, and two inverted repeats (IR) regions of 25,682 bp. The complete chloroplast genome contained a total of 130 genes, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes.
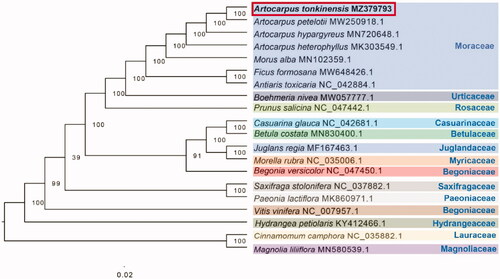
Complete chloroplast genomes of 19 other species were selected to confirm the phylogenetic position of A. tonkinensis, with Cinnamomum camphora and Magnolia liliiflora as outgroups. All of these complete chloroplast sequences were aligned by the MAFFT version 7.429 software (Katoh and Standley Citation2013) and trimmed by TrimAl (Capella-Gutierrez et al. Citation2009). A maximum-likelihood (ML) tree was inferred by ultrafast bootstrapping with 1000 replicates through IQ-TREE 1.6.12 (Nguyen et al. Citation2015) based on the TVM + F+R2 nucleotide substitution model, which was selected by ModelFinder (Kalyaanamoorthy et al. Citation2017). The findings of the phylogenetic analysis supported that Artocarpus belongs to the Moraceae family and proposed a sister relationship between Artocarpus and Morus ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank number MZ379793 (https://www.ncbi.nlm.nih.gov/nuccore/MZ379793.1/) and SRA number PRJNA738269 (https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA738269).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Orecchini E, Mondanelli G, Orabona C, Volpi C, Adorisio S, Calvitti M, Thuy TT, Delfino DV, Belladonna ML. 2020. Artocarpus tonkinensis extract inhibits LPS-triggered inflammation markers and suppresses RANKL-induced osteoclastogenesis in RAW264.7. Front Pharmacol. 11:593829.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLOS One. 7(2):e30619.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Thuy TT, Thien DD, Hung TQ, Tam NT, Anh NTH, Dung LK, Van Sung T, Delfino DV. 2017. Flavonol and proanthocyanidin glycosides from the leaves of Artocarpus tonkinensis. Chem Nat Compd. 53(4):759–753.
- Zhang XS, Wu ZY, Cao ZY. 1998. Moraceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 23. Beijing: Science Press; p. 55–56. [in Chinese]
- Zhou ZK, Michael GG. 2003. Moraceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 5. St. Louis: Science Press, Beijing & Missouri Botanical Garden Press; p. 21–73.