Abstract
Isoetes japonica A. Braun is a heterosporous quillwort living in low-altitude areas in Japan. In the present study, the complete chloroplast genome of I. japonica was assembled and annotated. This chloroplast genome is a circular structure of 145,517 bp in length, comprising a pair of inverted repeat (IR) regions of 13,204 bp each, a large single copy (LSC) region of 91,868 bp, and a small single copy (SSC) region of 27,241 bp. The chloroplast genome contains 135 genes, including 78 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. Phylogenetic analysis showed that I. japonica is sister to I. sinensis and I. yunguiensis. These results provide additional resources for the future studies on Isoetes species.
Keywords:
Isoetes japonica A. Braun (2n = 66) is a member of heterospores of Isoetes L., distributing in low-altitude areas of Japan (Takamiya Citation1999). I. japonica is either a submerged or emerged plant, growing in ponds, streams, and paddy fields (Takamiya et al. Citation1994). Megaspore ornamentation of this species present as reticulate and microspore surface is smooth (Takamiya Citation1999).
In the present study, leaf material was obtained from a specimen (voucher: Miyoshi Furuse 57289) collected from Pacang Town, Tochigi City, Tochigi Prefecture, Japan and deposited in China National Herbarium (PE, 01615847). The leaves were sent to Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China) for DNA extraction, and sequencing was performed on an Illumina HiSeq X Ten platform (Illumina, San Diego, CA, USA). The plastid genome was assembled using GetOrganelle v1.7.5 (Jin et al. Citation2018), and the results were viewed and edited by Bandage v0.8.1 (Wick et al. Citation2015). Assembled chloroplast genome was annotated by Geneious Prime 2021.0.3 (https://www.geneious.com) (Kearse et al. Citation2012) with I. engelmannii as the reference.
The complete plastid genome sequence of I. japonica (GenBank accession: MZ596344) is 145,517 bp in length, containing a large single-copy (LSC) region of 91,868 bp, a small single-copy (SSC) region of 27,241 bp, and a pair of inverted repeats (IRs) of 13,204 bp each. A total of 135 genes were annotated, including 78 protein-coding genes, 36 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. The overall GC content is 38.0%.
To figure out the phylogenetic position of I. japonica, a phylogenetic analysis was carried out with complete chloroplast genomes of 11 Isoetes species downloaded from GenBank, and two members of Lycopodiaceae (Lycopodium clavatum and Huperzia serrata) were set as outgroups. The sequences were aligned using MAFFT v7 in PhyloSuite v1.2.2 (Zhang et al. Citation2020). A maximum-likelihood (ML) tree was constructed by RAxML v8 with the GTRCAT nucleotide substitution model and 1,000 rapid bootstraps (Stamatakis Citation2014).
The ML tree indicated that I. nuttallii and I. cangae are two sole clades. I. hypsophila is sister to the clade of I. sinensis, I. yunguiensis, and I. japonica with a support value of 96%, and I. japonica was sister to I. sinensis and I. yunguiensis with a support value of 100% (). The above four species form a clade sister to the clade consisting of I. mattaponica, I. graniticola, I. engelmannii, I. melanospora, I. butleri, and I. valida, showing a 100% support value.
Figure 1. Maximum likelihood phylogenetic tree of 12 Isoetes species, with Lycopodium clavatum and Huperzia serrata as outgroups, constructed based on plastid genome sequences by RAxML. The number next to each node indicates the bootstrap value.
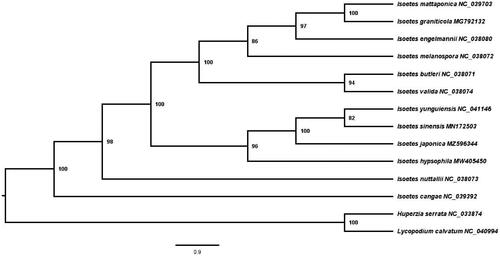
According to the ML tree, Isoetes japonica has close relationship to quillworts collected from China, this agree with the view Kim et al. (Citation2010) reported that its speciation may be associated with I. sinensis and I. taiwanensis but not supported by spore morphology features. Here, our result supports I. japonica is the sister clade with I. sinensis, we speculate it could possess another speciation pathway. Information presented in this paper and other related data on the net will assist in the future studies of Isoetes.
Acknowledgments
The authors would like to thank TopEdit (www.topeditsci.com) for its linguistic assistance during the preparation of this manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study can be obtained from GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ596344. The associated accession numbers of BioProject, SRA, and Bio-sample are PRJNA731595, SRR15196053, and SAMN20310797, respectively.
Additional information
Funding
References
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv.:256479.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kim C, Shin H, Chang YT, Choi HK. 2010. Speciation pathway of Isoetes (Isoetaceae) in East Asia inferred from molecular phylogenetic relationships. Am J Bot. 97(6):958–969.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Takamiya M. 1999. Natural history and taxonomy of Isoetes (Isoetaceae). Acta Phytotax Geobot. 50(1):101–138.
- Takamiya M, Watanabe M, Ono K. 1994. Biosystematic studies on the genus Isoetes in Japan I. Variations of the somatic chromosome numbers. J Plant Res. 107(3):289–297.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
