Abstract
Loropetalum chinense var. rubrum Yieh (L. chinense) is an evergreen shrub or small tree of Hamamelidaceae. In this study, the chloroplast genome sequence of L. chinense is 159,451 bp in length, consisting of a large single-copy region with 88,166 bp (LSC), a small single-copy region with 18,773 bp (SSC), and two inverted repeat regions with 26,256 bp (IRs). The GC content in the chloroplast genome of L. chinense is 38.0%. The chloroplast genome of L. chinense contained 125 genes, including 84 protein-coding genes, 37 tRNA genes, and 4 rRNA genes. The phylogenetic tree showed that L. chinense was closely related to L. subcordatum.
Loropetalum is a genus of Hamamelidaceae, including 4 species and 1 variety, mainly distributed in the subtropical regions of eastern Asia. L. chinense var. rubrum (L. chinense) is a member of the witch hazel family (Hamamelidaceae) and defined as a variety of L. chinense (Bao Citation2007). It has been reported that many kinds of unsaturated fatty acids existe in leaves of L. chinense, and its leaves could be used as Chinese medicine (Tang Citation2011). Our complete chloroplast genome data of L. chinense can contribute to a better understanding of the evolution of Loropetalum.
The fresh leaves of L. chinense were collected in Botanical Garden, Zhengzhou, China (34°44’51”N; 113°32’39”E). The voucher specimen was deposited at the Herbarium of Henan Agricultural University (voucher number:LC-20-0915). The total genomic DNA was extracted from fresh leaves of L. chinense using a modified CTAB method (Doyle and Doyle Citation1987). Sequencing was performed with the Illumina HiSeq2500 Platform (San Diego, CA). The raw reads were generated by Illumina paired-end sequencing after removing adapters. The low-quality sequences of raw reads used Fastp (https://github.com/OpenGene/Fastp) for quality control. Finally, the obtained clean reads were assembled by GetOrganelle pipeline v1.6.3a (https://github.com/Kinggerm/GetOrganelle) with the chloroplast genome of L. subcordatum (GenBank accession no. NC_037694.1) as the reference. The genome was automatically annotated by using the CpGAVAS2 pipeline (Shi et al. Citation2019), and start/stop codons and intron/exon boundaries were adjusted in Geneious 20.2.2 (https://www.geneious.com/).
The chloroplast genome sequence of L. chinense was submitted to NCBI, and the accession number was MW346664. The genome sequence of L. chinense is 159,451 bp in length, consisting of a large single-copy region with 88,166 bp (LSC), a small single-copy region with 18,773 bp (SSC), and two inverted repeat regions with 26,256 bp (IRs). The GC content in the chloroplast genome of L. chinense is 38.0%. The chloroplast genome of L. chinense contained 125 genes, including 84 protein-coding genes, 37 tRNA genes, and 4 rRNA genes.
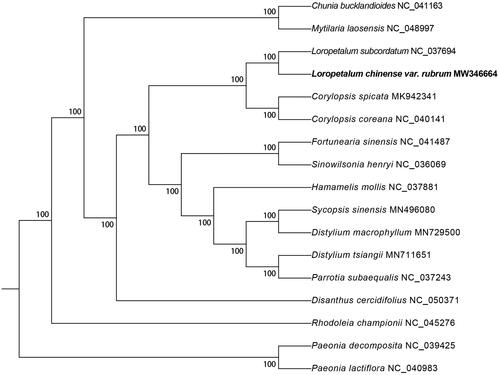
The phylogenetic tree was constructed in RAxML v8.2 (Stamatakis Citation2006) with 1000 bootstrap replicates. A total of 17 species were used and 2 Paeonia species as outgroup (Li et al. Citation2019). As shown in the phylogenetic tree (), the fifteen Hamamelidaceae species were organized into four clusters. The result strongly supported that L. chinense was closely related to L. subcordatum. This result was similar to the previous phylogenetic trees based on chloroplast genome sequences of Hamamelidaceae (Zhang et al. Citation2019).
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW346664. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA670186, SRA: SRS7961283, and SAMN17100623 respectively.
Additional information
Funding
References
- Bao Z, Chen B, Zhang H. 2007. Variation in morphological traits among Loropetalum chinense var. rubrum accessions. Horts. 42(2):399–402.
- Doyle JJ, Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Li H, Cheng X, Chen Y, Tan F. 2019. Complete plastome sequence of Rhodoleia championii Hook. f. (Hamamelidaceae). Mitochondrial DNA Part B. 4(2):3458–3459.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Stamatakis A. 2006. Raxml-vi-hpc: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Tang H, Zheng Q, Ge G, et al. 2011. Analysis of fatty acids in leaves of Loropetalum chinense and L. chinense var. rubrum by GC-MS. Chinese Medicinal Materials. 34(10):1549–1552.
- Zhang YY, Cai H, Dong J, Gong W, Li P, Wang Z. 2019. The complete chloroplast genome of Loropetalum subcordatum, a national key protected species in China. Conservation Genet Resour. 11(4):377–380.