Abstract
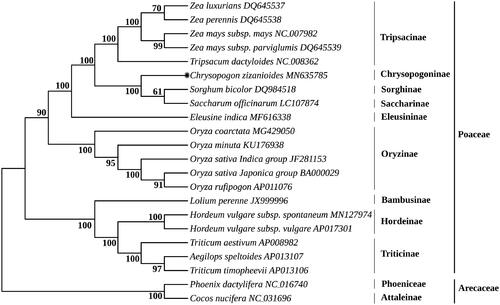
Vetiver grass (Chrysopogon zizanioides), is a perennial and tussock C4 grass from the genus Chrysopogon of Poaceae, which has been widely used as a natural and inexpensive resource for multifarious environmental applications. The complete mitogenome of C. zizanioides was 551,622 bp in length, containing 40 protein-coding genes (PCGs), 19 transfer RNA genes (tRNAs), and six ribosomal RNA genes (rRNAs). All PCGs started with ATG and stopped with TNN (TAA, TAG, and TGA). The overall nucleotide composition is: 28.2% A, 28.2% T, 21.7% G, and 21.9% C, with a biased A + T content of 56.4%. Phylogenetic analysis using 14 PCGs of 22 species showed that C. zizanioides display a close relationship with Saccharum officinarum (LC107874) and Sorghum bicolor (DQ984518) in Poaceae.
Introduction
Chrysopogon zizanioides (L.) Roberty, is a perennial and tussock C4 grass from the genus Chrysopogon of Poaceae, which has been widely cultivated in the tropics and sub-tropics of the world (Yaseen et al. Citation2014; Sigmon et al. Citation2017). C. zizanioides is a miracle grass that can tolerate extreme climatic and soil variations, which has been widely used as a natural and inexpensive resource for multifarious environmental applications, including conservation and detoxification of degraded soil and water and also for mitigation of flood and landslide disasters (Kemper Citation1993; Adams et al. Citation2004; Chakrabarty et al. Citation2015). To date, there have been more than 256 mitogenomes from land plants that are available, while no mitogenome was from the genus Chrysopogon (https://www.ncbi.nlm.nih.gov/genome/browse#!/organelles/, accessed 1 December 2020). Moreover, the narrow genetic and genomic resources of C. zizanioides also obviously limited our understanding of this important C4 grass at the genome level. In this study, we first determined the complete mitogenome sequences of C. zizanioides using Illumina next-generation sequencing technology, and performed an analysis of the phylogenetic relationships among other 21 species with the available mitogenome data deposited in GenBank.
Materials and methods
The fresh leaves of C. zizanioides ‘Xiangnan 1’ were collected from Nanyang City, Henan Province, China (112°52′E, 33°00′N). The specimen was deposited at the Collaborative Innovation Center of Water Security for Water Source Region of Mid-line of South-to-North Diversion Project of Henan Province under the voucher number NYNU_CN102986400A, Nanyang Normal University, China (Jibao Chen, [email protected]). Whole genome sequencing was conducted on Illumina HiSeq 3000 sequencing platform with 350 bp paired-end (Biomarker Technologies Corporation, Beijing, China). The mitogenome assembly of C. zizanioides was performed according to Lloyd Evans et al. (Citation2019) using the reference mitogenome dataset of S. officinarum (LC107874) and S. bicolor (DQ984518). All mitochondrial protein-coding genes (PCGs), transfer RNA (tRNA), and ribosomal RNA (rRNA) genes were initially annotated using Plann. PCGs were further predicted or modified using NCBI ORFfinder and annotated by BLASTP searches against the NCBI NR. tRNAs were also predicted or identified using tRNAscan-SE v2.0 (Bleasby and Wootton Citation1990; Huang and Cronk Citation2015; Chan and Lowe Citation2019). The phylogenetic tree was reconstructed using the Bayesian analysis (BI) and maximum-likelihood (ML) analyses. The best-fit model of nucleotide substitution for sequences with the Bayesian information criterion (BIC) was determined by jModelTest 2.0.2 (Darriba et al. Citation2012). The Bayesian phylogenetic analysis was performed using MrBayes 3.2.5 (Ronquist et al. Citation2012). ML analysis was conducted using RAxML with the GTR + G+I model (Stamatakis Citation2014). Bootstrap values were calculated using 1000 replicates to assess node support.
Results and discussion
The complete sequence size of C. zizanioides mitogenome was 551,622 bp in length (GenBank accession no. MN635785), which is well within the size range observed in the completely assembled other Poaceae mitogenomes. The overall nucleotide composition is: 28.2% A, 28.2% T, 21.7% G, and 21.9% C, with a biased A + T content of 56.4%. It presents a typical gene set of 40 PCGs, 19 tRNA genes, and six rRNA genes. All PCGs started with the conventional initiation codon of ATG, 22 of them used TAA, 10 PCGs (atp9, 2 ccmC, ccmFN, cob, mttB, matR, nad7, rps1, and rps3) used TAG, and eight PCGs (atp1, ccmB, ccmFC, cox3, nad4, rps2, rps12, and rps13) used TGA as the termination codon. All 19 tRNAs could fold into a classical cloverleaf structure except for trnY-GTA, trnS-GCT, and trnS-TGA, with their length ranging from 71 bp (trnC-GCA) to 91 bp (trnS-TGA). The lengths of rrn5, rrn18, and rrn26 were 118 bp, 1960 bp, and 3516 bp, including the rrn5 and rrn18 existed in a cluster.
The nucleotide sequences of 14 PCGs were used to understand the phylogenetic relationships of C. zizanioides with other 21 species using the ML method. Phylogenetic relationships obtained using the ML method were identical to those obtained using the BI analysis. The consensus topology of phylogenetic trees for 22 species extremely highly support that C. zizanioides, S. officinarum, and S. bicolor are a sister group ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MN635785. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA752393, SRR15371682, and SAMN20594899, respectively.
Additional information
Funding
References
- Adams RP, Habte M, Park S, Dafforn MR. 2004. Preliminary comparison of vetiver root essential oils from cleansed (bacteria- and fungus-free) versus non-cleansed (normal) vetiver plants. Biochem Syst Ecol. 32(12):1137–1144.
- Bleasby AJ, Wootton JC. 1990. Construction of validated, non-redundant composite protein sequence databases. Protein Eng. 3(3):153–159.
- Chakrabarty D, Chauhan PS, Chauhan AS, Indoliya Y, Lavania UC, Nautiyal CS. 2015. De novo assembly and characterization of root transcriptome in two distinct morphotypes of vetiver, Chrysopogon zizanioides (L.) Roberty. Sci Rep. 5:18630.
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Kemper WD. 1993. Vetiver grass: a thin green line against erosion. J Soil Water Conserv. 48(5):426.
- Lloyd Evans D, Hlongwane TT, Joshi SV, Riaño Pachón DM. 2019. The sugarcane mitochondrial genome: assembly, phylogenetics and transcriptomics. Peer J. 7:e7558.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Sigmon BA, Adams RP, Mower JP. 2017. Complete chloroplast genome sequencing of vetiver grass (Chrysopogon zizanioides) identifies markers that distinguish the non-fertile ‘Sunshine’ cultivar from other accessions. Ind Crop Prod. 108:629–635.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Yaseen M, Singh M, Ram D. 2014. Growth, yield and economics of vetiver (Vetiveria zizanioides L. Nash) under intercropping system. Ind Crop Prod. 61:417–421.