Abstract
Gynostemma microspermum C. Y. Wu et S. K. Chen is an endemic creeping herbaceous species mainly distributed in dense forests on limestone in northwestern China. Here, the complete chloroplast genome sequence of G. microspermum was obtained by Illumina pair-end sequencing. The circular complete chloroplast genome of G. microspermum is 158,692 bp in length and contains a large single copy region (87,452 bp), a small single copy region (19,068 bp) and two short inverted repeat regions (26,086 bp). The genome sequence encodes 133 genes including 87 protein-coding genes, 37 transfer RNA genes, 8 ribosomal RNA genes and 1 pseudogene. The maximum likelihood (ML) phylogeny estimation shows that G. microspermum is sister to all other analyzed species of the genus Gynostemma with high bootstrap support.
The genus Gynostemma Bl. belonging to family Cucurbitaceae consists of 17 creeping herbaceous species mainly distributed in east and southeast Asia. They are usually used as tea and considered as medical plants due to their anti-inflammatory properties, anticancer effects and weight controlling function (Xie et al. Citation2010). Gynostemma microspermum C. Y. Wu et S. K. Chen is an endemic species mainly distributed in dense forests on limestone in northwestern China at an altitude between 800 m and 1400 m (Chen Citation1995). Unfortunately, wild population of G. microspermum was severely decimated during the past years because of habitat destruction and urbanization, which could accelerate biodiversity declines and species extinctions (Ceballos et al. Citation2015). Therefore, more measures should be taken to protect the wild resources of G. microspermum urgently and ensure species diversity. Previous studies have shown that genetic and genomic researches would make contribution to species conservation. However, some studies focus on genetic relationships of Gynostemma species based on chloroplast genome or few gene fragments (Zhao et al. Citation2015; Zhang et al. Citation2017), but no genomic studies on G. microspermum. Thus, we assembled and characterized the complete chloroplast genome sequence of G. microspermum based on the Illumina pair-end sequencing. This study will provide a valuable complete chloroplast genomic resource and contribute to the further study on the phylogenetic analysis, systematic evolution and conservation genetics of G. microspermum.
Fresh and healthy leaves of G. microspermum were collected from adult plants in Mengla county (Yunnan, China; 21.74°N, 101.39°E), and a specimen was deposited at Northwest University (Xiao Zhang, [email protected]) under the voucher number NWU020161211. Total genomic DNA (number: DNA202008230024) was extracted by CTAB method (Doyle Citation1987) using for high-throughput sequencing with the Illumina Hiseq 2500 platform by Genesky Biotechnologies Inc. (Shanghai, China). A total of 1.3 Gb raw reads were obtained with an average length of 149.9 bp yielding 1232.4× coverage of the genome. After quality-trimmed using the CLC Genomics Workbench v7.5 (CLC bio, Aarhus, Denmark) program, reference-guided assembly was performed twice to construct the chloroplast genome with the program MITObim v1.7 (Hahn et al. Citation2013) using published Gynostemma pentaphyllum (KX014626) and Gynostemma cardiospermum (KX852299) (Zhang et al. Citation2017) as references, respectively. Software Geneious R8 (Biomatters Ltd, Auckland, New Zealand) was used to annotate the complete chloroplast genome. Finally, the annotated chloroplast genome sequence of G. microspermum has been submitted to the GenBank with the accession number MZ286581.
The complete chloroplast genome of G. microspermum is 158,692 bp in length and contains a large single copy region (LSC, 87,452 bp), a small single copy region (SSC, 19,068 bp), and a pair of inverted repeat regions (IRa and IRb, 26,086 bp). The genome sequence encodes 133 genes including 87 protein-coding genes (CDS), 37 transfer RNA (tRNA) genes, 8 ribosomal RNA (rRNA) genes and 1 pseudogene. Among these annotated genes, 16 (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps12, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) of them exist a single intron. Most of the genes occur in a single copy, but 8 protein-coding genes (ndhB, rpl2, rpl23, ycf1, ycf2, rps7, rps12, orf70), 7 tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC) and 4 rRNA genes (rrn16, rrn23, rrn4.5, rrn5) occur in twice in IRa and IRb. The gene infA, which is proved to be a translation-related gene, is identified to be a pseudogene.
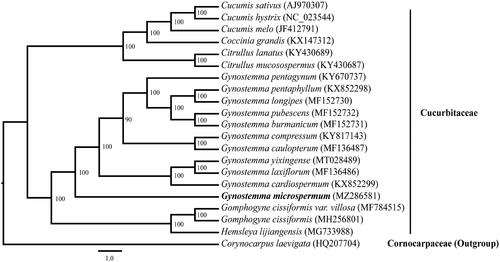
A total of 21 complete chloroplast genome sequences (Plader et al. Citation2007; Atherton et al. Citation2010; Rodriguez-Moreno et al. Citation2011; Sousa et al. Citation2016; Zhang et al. Citation2017, Citation2018a, Citation2018b) were selected to construct the phylogenetic relationships among the main representatives of Cucurbitales with Corynocarpus laevigata (HQ207704) as outgroup (). The maximum likelihood (ML) phylogenetic analysis was performed using RAxML v7.2.8 (Stamatakis Citation2006) performed with 1000 replicates. The ML phylogeny estimation shows that G. microspermum is sister to all other analyzed species of the genus Gynostemma with high bootstrap support. This study on the complete chloroplast genome of G. microspermum would provide information to the demonstration of chloroplast genome structure and the understanding of its evolution.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession number MZ286581. The raw sequence data used in this research were deposited successfully with registered numbers of associated BioProject, Bio-Sample and SRA: PRJNA733705, SAMN19433685, and SRR14689372, respectively.
Additional information
Funding
References
- Atherton RA, McComish BJ, Shepherd LD, Berry LA, Albert NW, Lockhart PJ. 2010. Whole genome sequencing of enriched chloroplast DNA using the Illumina GAII platform. Plant Methods. 6:22.
- Ceballos G, Ehrlich PR, Barnosky AD, García A, Pringle RM, Palmer TM. 2015. Accelerated modern human-induced species losses: entering the sixth mass extinction. Sci Adv. 1(5):e1400253.
- Chen S. 1995. A classificatory system and geographical distribution of the genus Gynostemma Bl. (Cucurbitaceae). Acta Phytotaxonomica Sinica. 33(4):403–410.
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Plader W, Yukawa Y, Sugiura M, Malepszy S. 2007. The complete structure of the cucumber (Cucumis sativus L.) chloroplast genome: its composition and comparative analysis. Cell Mol Biol Lett. 12 (4):584–594.
- Rodriguez-Moreno L, Gonzalez VM, Benjak A, Marti MC, Puigdomenech P, Aranda MA, Garcia-Mas J. 2011. Determination of the melon chloroplast and mitochondrial genome sequences reveals that the largest reported mitochondrial genome in plants contains a significant amount of DNA having a nuclear origin. BMC Genomics. 12:424.
- Sousa A, Bellot S, Fuchs J, Houben A, Renner S. 2016. Analysis of transposable elements and organellar DNA in male and female genomes of a species with a huge Y chromosome reveals distinct Y centromeres. Plant J. 88 (3):387–396.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Xie Z, Liu W, Huang H, Slavin M, Zhao Y, Whent M, Blackford J, Lutterodt H, Zhou H, Chen P, et al. 2010. Chemical composition of five commercial Gynostemma pentaphyllum samples and their radical scavenging, antiproliferative, and anti-inflammatory properties. J Agric Food Chem. 58(21):11243–11249.
- Zhang X, Li H, Zhou T, Yang Y, Zhao G. 2018a. The complete chloroplast genome of Gynostemma compressum. Conservation Genet Resour. 10(2):141–144.
- Zhang X, Zhou T, Kanwal N, Zhao Y, Bai G, Zhao G. 2017. Completion of eight Gynostemma BL. (Cucurbitaceae) chloroplast genomes: characterization, comparative analysis, and phylogenetic relationships. Front Plant Sci. 8:1583.
- Zhang X, Zhou T, Yang J, Sun J, Ju M, Zhao Y, Zhao G. 2018b. Comparative analyses of chloroplast genomes of cucurbitaceae lights into selective pressures and phylogenetic relationships. Molecules. 23 (9):2165.
- Zhao Y, Zhou T, Li Z, Zhao G. 2015. Characterization of global transcriptome using illumina paired-end sequencing and development of EST-SSR markers in two species of Gynostemma (Cucurbitaceae). Molecules. 20(12):21214–21231.