Abstract
Comaster schlegelii, belonging to the family Comatulidae, is a variable feather star distributed in the Pacific Ocean. The complete mitochondrial genome of this species was 15,887 bp in length, consisting of 13 protein-coding genes, 22 transport RNA genes, two ribosomal RNA genes and one control region. The whole mitochondrial genome of C. schlegelii had a high AT content of 72.73%. The phylogenetic relationship was reconstructed with 16 relevant echinoderms, which revealed C. schlegelii was closely clustered with Anneissia pinguis in the family Comatulidae.
Comaster schlegelii (Carpenter, 1881), variable bushy feather star with multicolor, is a crinoid in the order Comatulida, family Comatulidae. This species is common in the shallow-water regions, especially in the coral reefs of the western Pacific Ocean (Summers et al. Citation2017). In accordance with other feather stars, C. schlegelii is a suspension feeder and spreads out its arms and pinnules to feed. The phylogenetic relationships within Crinoidea are controversial and lack sufficient molecular evidence to validate. Till now, as for the progress of mitochondrial genome research, only nine species from four families were published, including the family Antedonidae (Nam et al. Citation2020), Sclerocrinidae (Scouras and Smith Citation2006), Comatulidae (Scouras and Smith Citation2006) and Mariametridae (Ma et al. Citation2019). For the family Comatulidae, only two mitochondrial genomes from the genus Anneissia (Kim and Shin 2021) and Phanogenia (Scouras and Smith Citation2006) were reported. Therefore, we sequenced the mitochondrial genome of C. schlegelii and enrich the comprehension of the genetic phylogenetic relationship of the Comatulidae family compared with other echinoderms.
The C. schlegelii sample was collected from Wuzhizhou Island in Sanya, China (18.31°N, 109.76°E, ≈ 12 m). The tissue sample was preserved in MNR Key Laboratory of Marine Eco-Environmental Science and Technology, First Institute of Oceanography, Ministry of Natural Resources (NO. FIO-ECH-HBH01). The genomic DNA of C. schlegelii was extracted using QIAamp Fast DNA Tissue Kit (QIAGEN, Germany). After quality control, the qualified DNA was sheared, processed to prepare 350 bp DNA library and pair-end sequenced on the Illumina Novaseq 5000 platform (PE150, 10×) in the Biomarker Corporation (Beijing, China).
The raw data were trimmed by Trimmomatic v0.38 (Bolger et al. Citation2014), and the filtered data were assembled using SPAdes v3.6.1 (Bankevich et al. Citation2012). Nucleotide Blast was performed with other species from Crinoidea in GenBank as the references to query the fragments of mitochondrial genome. Geseq (Tillich et al. Citation2017) and MITOs (Hahn et al. Citation2013) were used for preliminary annotation, and tRNA was annotated by tRNA scan-SE (Lowe and Chan Citation2016). The alignment and correction of amino acids were performed according to MEGA v7.0 software (Kumar et al. Citation2016) and the position of coding genes was corrected by Unipro UGENE (Okonechnikov et al. Citation2012). The complete mitochondrial genome was submitted to GenBank with accession number MW526391.
The complete mitochondrial genome for C. schlegelii was 15,887 bp in length, including 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and one control region. The overall base composition was 26.73% A, 11.38% C, 15.89% G, and 46.00% T. The gene order of C. schlegelii was consistent with other species from the Comatulidae, indicating that the gene order was conserved in this family.
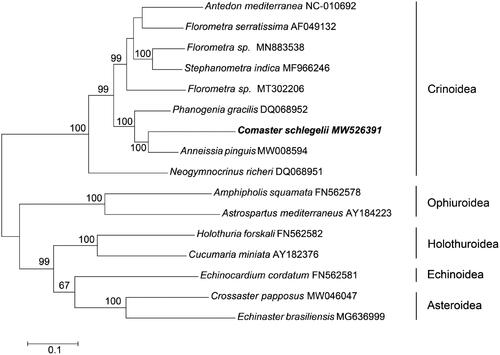
A Maximum Likelihood tree was constructed by MEGA v7.0 software (Kumar et al. Citation2016) using the dataset containing 13 PCGs and 2 rRNA. The nine published mitochondrial genomes from Crinoidea were chosen and the seven outgroups were from other classes. The phylogenetic tree depicted that C. schlegelii formed a branch with A. pinguis (), as the representative of the family Comatulidae. They clustered together with other feather stars to form the class Crinoidea. This mitochondrial genome analysis provides the foundation for phylogeny studies in the genus Comaster. Further studies call for more mitochondrial genomes resources to explore the relationship within Crinoidea.
Figure 1. Maximum-likelihood tree for the feather star Comaster schlegelii and GenBank representatives of the phylum Echinodermata. The tree was constructed using 13 protein-coding genes and 2 rRNA. The tree was based on the Kimura 2-parameter model of nucleotide substitution. The numbers at the nodes are bootstrap percent probability values based on 1000 replications. The mitochondrial genome sequence obtained at the present study was indicated in bold type.
Acknowledgements
Thanks to Jian Guo and Lei Zhao for DNA extraction, and thanks to Yixuan Li and Wenge Shi for giving suggestions on the modification of the article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov. The accession number of the complete mitochondrial genome is MW526391. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA692719, SUB8902528 and SAMN17367682, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19 (5):455–477.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads — a baiting and iterative mapping approach. Nucleic Acids Research. 41 (13):e129–e129.
- Kim P, Shin S. 2021. The complete mitochondrial genome of Anneissia pinguis (Crinoidea, Articulata, Comatulidae), from South Korea. Mitochondrial DNA B Resour. 6 (8):2337–2338.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33 (7):1870–1874.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44 (W1):W54–57.
- Ma S, Zhang H, Wang X, Yin J, Shen P, Lin Q. 2019. Characterization and phylogenetic analysis of the complete mitochondrial genome of Stephnometra indica (Pelmatozoa: Crinoidea). Mitochondrial DNA B Resour. 4 (2):2283–2284.
- Nam SE, Park HS, Rhee J-S. 2020. Characterization and phylogenetic analysis of the complete mitochondrial genome of Florometra species (Echinodermata, Crinoidea). Mitochondrial DNA Part B. 5 (2):2010–2011.
- Nam SE, Park HS, Rhee J-S. 2020. Complete mitochondrial genome of the Crinoid echinoderm, Florometra species (Echinodermata, Crinoidea). Mitochondrial DNA B Resour. 5 (1):852–853.
- Okonechnikov K, Golosova O, Fursov M, UGENE Team. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 28 (8):1166–1167.
- Scouras A, Smith MJ. 2006. The complete mitochondrial genomes of the sea lily Gymnocrinus richeri and the feather star Phanogenia gracilis: signature nucleotide bias and unique nad4L gene rearrangement within crinoids. Mol Phylogenet Evol. 39 (2):323–334.
- Summers MM, Messing CG, Rouse GW. 2017. The genera and species of Comatulidae (Comatulida: Crinoidea): taxonomic revisions and a molecular and morphological guide. Zootaxa. 4268 (2):151–190.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
