Abstract
In the present study, the complete mitochondrial genome of Aspergillus terricola É.J. Marchal 1893 was sequenced and assembled. The mitochondrial genome of A. terricola was composed of circular DNA molecules, with a total size of 28,689 bp. The GC content of the A. terricola mitochondrial genome was 26.34%. A total of 18 protein-coding genes (PCGs), 2 ribosomal RNA (rRNA) genes, and 26 transfer RNA (tRNA) genes were detected in the A. terricola mitochondrial genome. Phylogenetic analysis based on the combined mitochondrial gene dataset indicated that the A. terricola exhibited a close relationship with A. parasiticus.
Aspergillus is a highly diverse genus, widely distributed in environments, including soil, water, plant tissue and so on. Up to now, hundreds of species have been described in the genus Aspergillus. More and more attention has been paid to the research of Aspergillus, which plays an important role in agriculture, industry and food research (Facchini et al. Citation2012; Michelin et al. Citation2012). Aspergillus terricola É.J. Marchal 1893 has been isolated from fermented products (Facchini et al. Citation2012). It can produce high activity protease, including acidic, neutral and alkaline protease, and has been paid more and more attention in the food industry (Yadav et al. Citation2009; Michelin et al. Citation2010; Singh et al. Citation2010; Michelin et al. Citation2011). The mitochondrial gene is an important molecular marker to analyze the phylogenetic relationship and evolution of species (Zhang et al. Citation2017; Wu et al. Citation2021; Q Li, L Li, et al. Citation2021). However, up to now, the mitochondrial genome characteristics of A. terricola have not been understood. This study served as the first report on the complete mitochondrial genome of A. terricola, which will promote the understanding of phylogeny and evolution of this important fungal species.
The specimen (A. terricola) was collected from Luzhou, Sichuan, China (105.40 E; 28.91 N) from the soy sauce fermentation system. A specimen was deposited in the collection center of Luzhou Vocational and Technical College (Yue Deng, [email protected]) under the voucher number L03. The complete mitochondrial genome of A. terricola was sequenced and assembled according to previously described methods (Li et al. Citation2019; Wang, Song, et al. Citation2020; X Wang, YJ Wang, et al. Citation2020). In brief, genomic DNA of A. terricola was extracted using a fungal DNA kit (Cat. #D3390-00, Omega Bio-Tek, Norcross, GA, USA) according to the manufacturer’s instructions. NEBNext Ultra II DNA Library Prep Kits (NEB, Beijing, China) were used to construct sequencing libraries. Whole genomic sequencing was performed by the Illumina HiSeq 2500 Platform (Illumina, San Diego, CA, USA). We conducted quality control steps to obtain clean reads from the raw sequencing reads according to previous studies (Li, Yang, et al. Citation2020). The complete mitochondrial genome of A. terricola was assembled using NOVOPlasty v4.3.1 with the obtained clean reads under the k-mer size of 27 (Dierckxsens et al. Citation2017; Li, Ren, et al. Citation2020). The complete mitochondrial genome of A. terricola was annotated using MITOS (Bernt et al. Citation2013) and MFannot (Valach et al. Citation2014) based on the genetic code 4. PCGs or ORFs in the A. terricola mitochondrial genome were predicted using the NCBI Open Reading Frame (ORF) Finder (NCBI Resource Coordinators Citation2018) and annotated by BLASTP searches against the NCBI non-redundant protein sequence database (Bleasby and Wootton Citation1990). We then predicted the tRNA genes in the A. terricola mitochondrial genome using tRNAscan-SE v1.3.1 (Lowe and Chan Citation2016).
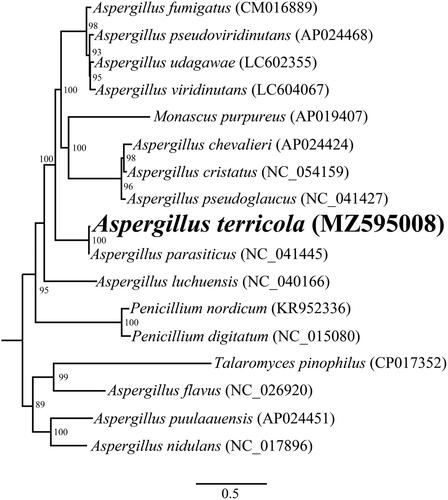
The complete mitochondrial genome of A. terricola is 28,689 bp in length. The base composition of the A. terricola mitochondrial genome is as follows: A (35.82%), T (37.83%), G (14.55%) and C (11.80%). A total of 18 PCGs (cox1, cox2, cox3, cob, atp6, atp8, atp9, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, rps3, orf205, orf153, and orf650), 2 ribosomal RNA genes (rns and rnl), and 26 transfer RNA genes were detected in the A. terricola mitochondrial genome. The mitochondrial genome of A. terricola contained 3 introns, 2 of which were located in cox1and one was located in the rnl gene. Two introns belong to an unknown type, and the other belongs to group I. We named these introns according to the previously described method (S Zhang and YJ Zhang Citation2019). To reveal the phylogenetic relationships of the Eurotiales species, a phylogenetic tree for 17 Eurotiales species was constructed. Single mitochondrial genes were first aligned by using MAFFT v7.037 (Katoh et al. Citation2019), and then concatenated into a pseudogene dataset by using SequenceMatrix v1.7.8 (Vaidya et al. Citation2011). We detected best-fit models of evolution and partitioning schemes using PartitionFinder 2.1.1 (Lanfear et al. Citation2017). The phylogenetic relationships of the 17 Eurotiales species were analyzed by using RAxML v 8.0.0 (Stamatakis Citation2014). The maximum likelihood method (ML) method was used to construct the phylogenetic tree based on the combined 14 core PCGs (Cheng et al. Citation2021; Li, Wu, et al. Citation2021). We assessed bootstrap values (BS) through an ultrafast bootstrap approach, with 10,00 replicates. As shown in the phylogenetic tree (), the mitochondrial genome of A. terricola exhibited a close relationship with A. parasiticus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ595008. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA747851, SRR15184095, and SAMN20297433, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bleasby AJ, Wootton JC. 1990. Construction of validated, non-redundant composite protein sequence databases. Protein Eng. 3(3):153–159.
- Cheng J, Luo Q, Ren YH, Luo Z, Liao WL, Wang X, Li Q. 2021. Panorama of intron dynamics and gene rearrangements in the phylum Basidiomycota as revealed by the complete mitochondrial genome of Turbinellus floccosus. Appl Microbiol Biotechnol. 105(5):2017–2032.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Facchini FDA, Reis VRA, Roth AP, Magalhaes KA, Peixoto-Nogueira SC, Casagrande DR, Reis RA, Polizeli MDTM. 2012. Effects of Aspergillus spp. exogenous fibrolytic enzymes on in vitro fermentation of tropical forages. J Sci Food Agric. 92(12):2569–2573.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Li Q, Li L, Feng H, Tu W, Bao Z, Xiong C, Wang X, Qing Y, Huang W. 2021. Characterization of the complete mitochondrial genome of Basidiomycete yeast Hannaella oryzae: intron evolution, gene rearrangement, and its phylogeny. Front Microbiol. 12:646567.
- Li Q, Ren Y, Shi X, Peng L, Zhao J, Song Y, Zhao G. 2019. Comparative mitochondrial genome analysis of two ectomycorrhizal fungi (Rhizopogon) reveals dynamic changes of intron and phylogenetic relationships of the subphylum Agaricomycotina. IJMS. 20(20):5167.
- Li Q, Ren Y, Xiang D, Shi X, Zhao J, Peng L, Zhao G. 2020. Comparative mitogenome analysis of two ectomycorrhizal fungi (Paxillus) reveals gene rearrangement, intron dynamics, and phylogeny of basidiomycetes. IMA Fungus. 11(1):12.
- Li Q, Wu P, Li L, Feng H, Tu W, Bao Z, Xiong C, Gui M, Huang W. 2021. The first eleven mitochondrial genomes from the ectomycorrhizal fungal genus (Boletus) reveal intron loss and gene rearrangement. Int J Biol Macromol. 172:560–572.
- Li Q, Yang L, Xiang D, Wan Y, Wu Q, Huang W, Zhao G. 2020. The complete mitochondrial genomes of two model ectomycorrhizal fungi (Laccaria): features, intron dynamics and phylogenetic implications. Int J Biol Macromol. 145:974–984.
- Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–7.
- Michelin M, Peixoto-Nogueira SC, Betini JHA, da Silva TM, Jorge JA, Terenzi HF, Polizeli MLTM. 2010. Production and properties of xylanases from Aspergillus terricola Marchal and Aspergillus ochraceus and their use in cellulose pulp bleaching. Bioprocess Biosyst Eng. 33(7):813–821.
- Michelin M, Polizeli MDTD, da Silva DP, Ruzene DS, Vicente AA, Jorge JA, Terenzi HF, Teixeira JA. 2011. Production of xylanolytic enzymes by Aspergillus terricola in stirred tank and airlift tower loop bioreactors. J Ind Microbiol Biotechnol. 38(12):1979–1984.
- Michelin M, Polizeli MDTM, Ruzene DS, Silva DP, Ruiz HA, Vicente AA, Jorge JA, Terenzi HF, Teixeira JA. 2012. Production of xylanase and β-xylosidase from autohydrolysis liquor of corncob using two fungal strains . Bioprocess Biosyst Eng. 35(7):1185–1192.
- NCBI Resource Coordinators. 2018. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 46(D1):D8–D13.
- Singh RS, Bhari R, Kaur HP, Vig M. 2010. Purification and characterization of a novel thermostable mycelial lectin from Aspergillus terricola. Appl Biochem Biotechnol. 162(5):1339–1349.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Vaidya G, Lohman DL, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 27(2):171–180.
- Valach M, Burger G, Gray MW, Lang BF. 2014. Widespread occurrence of organelle genome-encoded 5S rRNAs including permuted molecules. Nucleic Acids Res. 42(22):13764–13777.
- Wang X, Song A, Wang F, Chen M, Li X, Li Q, Liu N. 2020. The 206 kbp mitochondrial genome of Phanerochaete carnosa reveals dynamics of introns, accumulation of repeat sequences and plasmid-derived genes. Int J Biol Macromol. 162:209–219.
- Wang X, Wang YJ, Yao W, Shen JW, Chen MY, Gao M, Ren JN, Li Q, Liu N. 2020. The 256 kb mitochondrial genome of Clavaria fumosa is the largest among phylum Basidiomycota and is rich in introns and intronic ORFs. IMA Fungus. 11(1):26. doi: https://doi.org/10.1186/S43008-020-00047-7
- Wu P, Bao Z, Tu W, Li L, Xiong C, Jin X, Li P, Gui M, Huang W, Li Q. 2021. The mitogenomes of two saprophytic Boletales species (Coniophora) reveals intron dynamics and accumulation of plasmid-derived and non-conserved genes. Comput Struct Biotechnol J. 19:401–414.
- Yadav S, Yadav PK, Yadav D, Yadav KDS. 2009. Purification and characterization of pectin lyase produced by Aspergillus terricola and its application in retting of natural fibers. Appl Biochem Biotechnol. 159(1):270–283.
- Zhang S, Zhang YJ. 2019. Proposal of a new nomenclature for introns in protein-coding genes in fungal mitogenomes. IMA Fungus. 10(15):15.
- Zhang YJ, Zhang HY, Liu XZ, Zhang S. 2017. Mitochondrial genome of the nematode endoparasitic fungus Hirsutella vermicola reveals a high level of synteny in the family Ophiocordycipitaceae. Appl Microbiol Biotechnol. 101(8):3295–3304.