Abstract
The short-faced mole (Scaptochirus moschatus) is a unique Chinese mammal that lives in burrows for life. In this study, we used Illumina NovaSeq sequencing to obtain the complete mitochondrial genome of the short-faced mole. The total length of the genome is 16,699 bp, containing 13 protein-coding genes, 22 transfer RNA genes (tRNA), 2 ribosomal RNA genes (rRNA), and 1 control region, with a base composition of 33.82% A, 26.89% T, 25.27% C, and 14.01% G. Phylogenetic analysis of the Talpidae by using complete mitochondrial genome sequences of 14 Talpidae species shows that short-faced mole is closely related to Parascaptor leucura.
Short-faced mole (Scaptochirus moschatus Milne-Edwards, 1867), the only representative species of genus Scaptochirus of the Talpidae, is a small mammal that lives in burrows for life, feeds on plant rhizomes and soil insects, etc (Smith and Johnston Citation2016). It has developed morphological and physiological characteristics adapted to burrowing, such as eye degeneration, fore claw valgus, smell and hearing sensitivity, etc (Sansalone et al. Citation2019; Chen et al. Citation2021). Several studies have focused on the phylogenetic and morphological evolution of Talpidae animals (He et al. Citation2014; Chen et al. Citation2017; He et al. Citation2017). ; A few mitochondrial genomes of Talpidae species have been reported, such as Iberian mole (Talpa occidentalis) (Gutiérrez et al. Citation2018), white-tailed mole (Parascaptor leucura) (Xie et al. Citation2021). However, there is no report on the mitochondrial genome of the short-faced mole.
In this study, short-faced mole was collected in Guanxian, Shandong Province, China (Latitude: 36°30′57″ N, Longitude: 115°25′24″ E; H: 43 m). The specimen was deposited at Qufu Normal University (Lei Chen, email: [email protected]) under the voucher number SMGF09. E.Z.N.A® Tissue DNA kit (OMEGA) was used to extract the total DNA of short-faced mole. The library was built by TruSeq™ Nano DNA Sample Prep Kit (Illumina), and the DNA was ultrasonically interrupted to 300–500bp by Covaris M220. After add adenine to the 3′ end, and connect the index adapter; the library was enriched and amplification by PCR. The PCR products were recycled by using a 2% agarose gel, quantified by TBS380 (Picogreen) and mixed according to the data proportions. Bridge PCR amplification was performed on the cBot solid phase carrier to generate clusters. The Illumina NovaSeq platform was used for sequencing with the pair end 150 bp protocol. After quality control of the raw data by using Trimmomatic (v0.39), clean data was assembled by using SPAdes (v3.14.1). Clean data with high coverage depth and long assembly length were selected as candidate sequences, and compared to the NT library to confirm the mitochondrial Scaffold sequence. The starting location and direction of the mitochondrial assembly sequence were determined according to the reference genome. MITOS software was used to predict the coding protein, tRNA and rRNA genes of the mitochondrial genome (Bernt et al. Citation2013). The start and stop codon positions of the genes were manually corrected to obtain a highly accurate conservative genome.
The complete mitochondrial genome sequence of short-faced mole was submitted to NCBI GenBank with the accession number MZ594566. It is 16,699 bp long, containing 13 protein-coding genes, 22 transfer RNA genes (tRNA), 2 ribosomal RNA genes (rRNA) and 1 control region. The total length of the 13 coding genes is 11,397 bp. The initiation codons of protein-coding genes are ATG, except for two protein coding genes (ND2, ND5, ) with the initiation codons of ATA, and one protein coding gene (ND3) initiates with ATT. The termination codons of seven protein coding genes ( COX1, COX2, ATP8, ATP6, ND4L, ND5, ND6) are TAA, while termination codons of five protein-coding genes (ND1,ND2, ND3, COX3, ND4) are incomplete. In addition, the termination codon of cytochrome b (Cyt b) is AGA, which is roughly consistent with the results of the white-tailed mole (Xie et al. Citation2021). GC content of short-faced mole mitochondrial genome is 39.28% and individual base proportions amounts to 33.82% A, 26.89% T, 25.27% C, and 14.01% G.
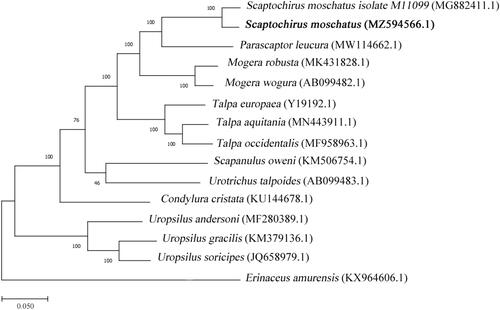
In order to explore the evolutionary status of the short-faced mole, we used the complete mitochondrial genome sequences of short-faced mole and 13 other species of the Talpidae for phylogenetic analysis. Erinaceus amurensis (KX964606.1) was used as the outgroup (Tsuchiya et al. Citation2000). MEGA X software is used to construct a phylogenetic tree under Tamura–Nei and maximum-likelihood model with 1000 bootstrap replication (Kumar et al. Citation2018) (). The result shows that short-faced mole is closely related to Parascaptor leucura, which is similar to the previous studies (He et al. Citation2017; Xie et al. Citation2021).
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study is openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession number MZ594566.1. The associated BioProject, SRA, and BioSample numbers are PRJNA769832, SRR16494732 and SAMN22419676, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69 (2):313–319.
- Chen L, Xu D, Zhu J, Wang S, Liu M, Sun MY, Wang GY, Song LY, Liu XY, Xie TY. 2021. Habitat environmental factors influence intestinal microbial diversity of the short-faced moles (Scaptochirus moschata). AMB Express. 11(1):93.
- Chen Z, He K, Huang C, Tao W, Lin L, Liu S, Jiang X. 2017. Integrative systematic analyses of the genus Chodsigoa (Mammalia: Eulipotyphla: Soricidae), with descriptions of new species. Zool J Linn Soc. 180(3):1–20.
- Gutiérrez J, Lamelas L, Aleix-Mata G, Arroyo M, Marchal JA, Palomeque T, Lorite P, Sánchez A. 2018. Complete mitochondrial genome of the Iberian mole Talpa occidentalis (Talpidae, Insectivora) and comparison with Talpa europaea. Genetica. 146(4-5):415–423.
- He K, Shinohara A, Helgen KM, Springer MS, Jiang X, Campbell KL. 2017. Talpid mole phylogeny unites shrew moles and illuminates overlooked cryptic species diversity. Mol Biol Evol. 34 (1):78–87.
- He K, Shinohara A, Jiang X, Campbell KL. 2014. Multilocus phylogeny of Talpine moles (Talpini, Talpidae, Eulipotyphla) and its implications for systematics. Mol Phylogenet Evol. 70:513–521.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 35 (6):1547–1549.
- Sansalone G, Colangelo P, Loy A, Raia P, Wroe S, Piras P. 2019. Impact of transition to a subterranean lifestyle on morphological disparity and integration in talpid moles (Mammalia, Talpidae). BMC Evol Biol. 19(1):179.
- Smith AT, Johnston CH. 2016. Scaptochirus moschatus (errata version published in 2017). The IUCN Red List of Threatened Species 2016: e.T41476A115188844.
- Tsuchiya K, Suzuki H, Shinohara A, Harada M, Wakana S, Sakaizumi M, Han SH, Lin K, Kryukov AP. 2000. Molecular phylogeny of East Asian moles inferred from the sequence variation of the mitochondrial cytochrome b gene. Genes Genet Syst. 75(1):17–24.
- Xie F, Chen D, Qin B, Fu C, Chen S. 2021. The complete mitochondrial genome of white-tailed mole (Parascaptor leucura). Mitochondrial DNA B Resour. 6(3):1112–1113.