Abstract
Vandenboschia striata is a common and widespread filmy fern of Hymenophyllaceae. Its complete chloroplast genome is 147,014 bp in length, including a large single copy (LSC) region of 89,886 bp, a small single copy (SSC) region of 20,850 bp, and a pair of inverted repeats (IRs) of 18,139 bp. Totally, 132 genes are predicted in the cp genome embodies, including 88 protein coding genes, 36 tRNA genes, and eight rRNA genes. A maximum-likelihood tree was constructed to explore phylogenetic relationship. The result showed that V. striata was sister to V.speciosa with 100% bootstrap support. The complete chloroplast genome sequences of V. striata would be beneficial to further phylogenetic survey on classification of the related species or genera in Hymenophyllaceae.
Vandenboschia striata (D. Don) Ebihara, well-known as misapplied name V. radicans in China, is a filmy fern belonging to Hymenophyllaceae. The fern is only 15–40 cm tall with a deep smoke-colored rhizome. Stipes are light brown and dark green-brown veins with dichotomous raised on each surface (Liu et al. Citation2013). As a common and widespread filmy fern, V. striata distributes in China (Guangdong, Guangxi, Guizhou, Hainan, Sichuan, Taiwan, and Yunnan), Bjutan, India, Japan, Laos, Myanmar, Nepal, and Vietnam (Liu et al. Citation2013). The plant prefers to grow on wet rocks near streams and slopes with altitudes 400–2700 m. Currently, the genera classification of Hymenophyllaceae, especially Vandenboschia, is greatly controversial (PPG I Citation2016). In addition, there are also problems involving in fern complex and hybridization such as V. radicans complex (Ebihara et al. Citation2005; Ebihara et al. Citation2009). Hence, sequencing of the complete chloroplast genome of V. striata will greatly contribute to solving these issues.
Vandenboschia striata was collected from Fairy Lake Botanical Garden, Shenzhen (22°35′8.39″N, 114°11′6.68″E), which was immediately treated with liquid nitrogen, and stored in the −80 °C. The specimen is deposited at the Herbarium of Sun Yat-sen University (Wang Ruonan: [email protected] and voucher number: Wang202101). We extracted the genomic DNA from approximate 100 mg fresh and young leaves using Tiangen Plant Genomic DNA Kit (Tiangen Biotech Co., Beijing, China), which was further fragmented into 300 bp. After the Illumina library was constructed, sequencing was performed on a Hiseq 2500 platform (Illumina Inc., San Diego, CA). About 2.93 Gb paired-end data were generated and 2.67 Gb high-quality clean data were further assembled using GetOrganelle (Jin et al. Citation2020). Protein coding genes, rRNA genes, and tRNA genes were separately annotated through PGA (Qu et al. Citation2019) and tRNAscan-SE programs (Lowe and Eddy Citation1997), followed by adjustment and confirmation of codon and intron/exon boundaries using Geneious v8.1 (Kearse et al. Citation2012). Upload GenBank number was MZ911852. Raw reads were deposited in the GenBank Sequence Read Archive (SRA: SRR15564830).
The complete chloroplast genome of V. striata is 147,014 bp in length, with an overall GC content of 37.5%. It demonstrates a typical quadripartite circular structure, involving in a large single copy (LSC) region of 89,886 bp, a small single copy (SSC) region of 20,850 bp, and a pair of inverted repeats (IRs) of 18,139 bp. Totally, 132 genes are predicted in the cp genome embodies, including 88 protein-coding genes, 36 tRNA genes, and eight rRNA genes. Three genes (clpP, rps12, and ycf3) contain two introns, and 11 genes (atpF, ndhA, ndhB, rpl2, rpoC1, rps16, trnA-UGC, trnI-GAU, trnG-UCC, trnL-CAA, and trnV-UAC) have one intron, whereas 13 genes (psbA, rps7, rps12, trnA-GUU, trnI-GAU, trnL-GAU, trnN-GUU, trnR-ACG, trnV-GAC, 4.5S rRNA, 5S rRNA, 16S rRNA, and 23S rRNA) have two copies.
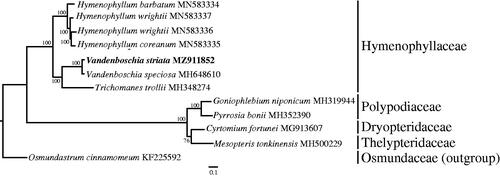
To investigate the phylogenetic relationship of V. striata, we downloaded 11 fern cp genomes from the NCBI database. These complete chloroplast genome sequences were aligned using MAFFT with default parameter (Katoh and Standley Citation2013), then a maximum-likelihood tree was constructed using RAxML v.8.2.12 with Osmundastrum cinnamomeum (KF225592) as outgroup and 1000 replicates (Stamatakis Citation2014). As a result, the phylogenetic tree showed that V. striata was sister to V. speciosa (). The complete chloroplast genome sequences of V. striata would be beneficial to the further phylogenetic survey on the classification of the related species or genera in Hymenophyllaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MZ911852, GenBank accession number MZ911852. Raw sequencing reads in this study are deposited in https://www.ncbi.nlm.nih.gov/sra/PRJNA756937, with SRA number SRR15564830.
Additional information
Funding
References
- Ebihara A, Ishikawa H, Matsumoto S, Lin SJ, Iwatsuki K, Takamiya M, Watano Y, Ito M. 2005. Nuclear DNA, chloroplast DNA, and ploidy analysis clarified biological complexity of the Vandenboschia radicans complex (Hymenophyllaceae) in Japan and adjacent areas. Am J Bot. 92(9):1535–1547.
- Ebihara A, Matsumoto S, Ito M. 2009. Hybridization involving independent gametophytes in the Vandenboschia radicans complex (Hymenophyllaceae): a new perspective on the distribution of fern hybrids. Mol Ecol. 18(23):4904–4911.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Liu JX, Zhang QY, Ebihara A, Iwatsuki K. 2013. Hymenophyllaceae. In: Wu ZY, Raven PH, Hong DY, eds. Flora of China. Vol. 2–3 (Pteridophytes). Beijing; St. Louis: Science Press; Missouri Botanical Garden Press; p. 93–109.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- PPG I. 2016. A community-derived classification for extant lycophytes and ferns. J Syst E. 54:563–603.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50–61.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.