Abstract
In this study, we sequenced the first complete chloroplast (cp) genome of a Peruvian chili pepper landrace, “arnacucho” (Capsicum chinense). This cp genome has a 156,931 bp in length with typical quadripartite structure, containing a large single copy (LSC) region (87,325 bp) and a 17,912 bp small single-copy (SSC) region, separated by two inverted repeat (IR) regions (25,847 bp); and the percentage of GC content was 37.71%. Arnaucho chili pepper chloroplast genome possesses 133 genes that consists of 86 protein-coding genes, 37 tRNA, eight rRNA, and two pseudogenes. Phylogenetic analysis revealed that this Peruvian chili pepper landrace is closely related to the undomesticated species C. galapagoense; all belong to the Capsiceae tribe.
Peru harbors the largest morphological diversity of cultivated chili peppers worldwide (Meckelmann et al. Citation2013), and they play a crucial role in the Peruvian cuisine and cultural traditions (Morales-Soriano et al. Citation2018). Arnaucho chili pepper is a landrace cultivated by small farmers in a restricted area named ‘Valle de Supe’ (around 200 km northern Lima) and is an important representative in the gastronomy of Lima region. This landrace possesses an annular constriction of the calyx, two flowers per axil, red ripe fruit color, triangular fruit shape, pointed fruit apex shape, greenish-yellow corolla (Aliaga et al. Citation2019). Even though NGS techniques are being employed to study Capsicum L. spp. genomes, very little is known about the genomics of Peruvian chili pepper. In addition, knowledge about Peruvian Capsicum chinense Jacquin 1777 phylogenetic relationships is scarce. Therefore, we here report the first complete chloroplast genome (cp) of a Peruvian chili pepper landrace by next-generation sequencing technology. Moreover, a phylogenetic tree of this species and its relatives is presented.
We collected young fresh leaves of arnaucho chili pepper from the Lima region (−10.8099889, −77.6953950). This specimen was deposited at the Germplasm Bank of INIA (https://www.gob.pe/inia, [email protected]) under the voucher number PER1002642. Total genomic DNA was extracted by CTAB method (Doyle and Doyle Citation1990). Pair-end clean reads were obtained by PE 150 library and the Illumina HiSeq 2500 platform. Adapters and low-quality reads were removed using Trim Galore (Martin Citation2011). We assembled the chloroplast genome using the GetOrganelle v1.7.2 pipeline (Jin et al Citation2020). Chloroplast genome was annotated with GeSeq in CHLOROBOX web service (Tillich et al Citation2017).
The total length of the chloroplast genome is 156,931 bp, which is 1,635 bp longer than one of the most economically important species in the Solanaceae family, potato (Solanum tuberosum). This cp genome presents a typical quadripartite structure, containing 87,325 bp as large single copy (LSC) region and 17,912 bp as small single-copy (SSC) region, separated by two inverted repeats (IR) regions (25,847 bp). The total GC content was 37.71%. Arnaucho chili pepper chloroplast genome contains 133 genes, including 86 protein-coding genes, 37 tRNA genes, 8 rRNA genes, and 2 pseudogenes. Similarly, chloroplast genome of the other two C. chinense (KX913217, KU041709) and C. galapagoense Hunziker 1956 (NC_033524) presents 86 protein-coding genes, 45 non-coding regions and 2 pseudogenes Most of these genes did not contain an intron; only 25 genes harbored one intron, and two genes (pafI, clpP1) contained two introns. Most genes occurred as a single copy, except 18 genes that were duplicated in IR regions. On the other hand, D’Agostino et al. (Citation2018) indicated that the plastome size of 11 Capsicum genotypes ranged from 156,836 bp in C. frutescens Linaeus 1753 to 157,390 in C. pubescens Ruiz & Pavon 1799. Moreover, they mentioned those plastomes contained 113 unique genes, 79 protein-coding, 4 rRNA, and 30 tRNA genes. The chloroplast genome sequence and annotation were submitted to NCBI with accession number MZ379791.
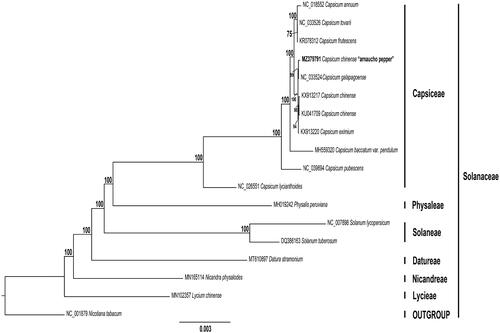
We constructed a maximum likelihood (ML) phylogenetic tree of 17 genomes obtained from GenBank. Each genome was aligned by MAFFT v7.475 (Katoh and Standley Citation2013). Then, we used GTR + GAMMA model of evolution to obtain the best-scoring ML tree, and then 1,000 nonparametric bootstrap inferences were performed with RAxML v8.2.11 (Stamatakis Citation2014). The phylogenomic analyses were consistent with a previous study that also employed plastome sequences (D’Agostino et al. Citation2018; Sebastin et al. Citation2019). Interestingly, ML phylogenetic analysis showed that arnacucho chili pepper is sister to C. galapagoense and sister to them is a clade containing C. chinense and C. eximium Hunziker 1950 (). This chili pepper landrace is considered a C. chinense species by many authors based mainly on morphological characters (Aliaga et al. Citation2019). However, its current taxonomy is questioned based on the present work. Carrizo García et al. (Citation2016) showed that C. galapagoense is nested among the closely related C. frutescens, C. chinense, and C. annuum Linaeus 1753 as demonstrated also by Ince et al. (Citation2010). In a more recent study, Shiragaki et al. (Citation2020) indicated that the C. chinense clade might be divided into two groups. Similarly, Tripodi et al. (Citation2021) demonstrated that C. chinense species grouped in two clusters.
Figure 1. Maximum likelihood reconstruction of the 18 (including arnacucho chili pepper, C. chinense) whole chloroplast genome sequences, and one outgroup. Numbers above the branches represent bootstrap values, with only values higher than 70% shown. Names given to clades refer to the tribes in Solanaceae. Branch length (number of substitutions per site) is represented by a scale bar (bottom).
To our best knowledge, this is the first report of a cp genome of a Peruvian chili pepper landrace. However, we consider that further studies using additional collections of Capsicum from a wider geographic area and examination of relevant type material are needed to provide a better understanding of taxonomic variation and nomenclature in the Capsicum clade. A similar process was followed by Arbizu, Ellison, et al. (Citation2016); Arbizu, Simon, et al. (Citation2016) and Martínez-Flores et al. (Citation2016) solving the species boundaries in another problematical group in Daucus. Our next step is to continue developing additional molecular tools for the arnaucho chili pepper that may shed light on elucidating its evolutionary history and promoting its adequate sustainable management, conservation and modern breeding.
Ethical approval
Research and collection of plant material was conducted according to the guidelines provided by INIA. Permission was granted by grower of “arnaucho” chili pepper to carry out research on this landrace.
Author contributions
Conceptualization, C.I.A.; methodology, C.I.A., C.L.S., R.D.F-M., J.C.Ch-G., and J.H.; formal analysis, C.I.A., C.L.S., and J.C.Ch-G.; resources, S.C-L., J.C.G-A., and J.L.M.; data curation, C.I.A.; writing—original draft preparation, C.I.A., C.L.S., S.C.-L.; writing—review and editing, C.I.A., C.L.S., S.C.-L., and J.C.G-A.; supervision, C.I.A., J.C.G-A., J.L.M.; funding acquisition, C.I.A., and J.L.M. All authors have read and agreed to the published version of the manuscript.
Acknowledgement
We thank Mr. Jorge Biondi for providing technical assistance in the laboratory.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that supports this study is openly available in Genbank of NCBI under the accession number MZ379791 (https://www.ncbi.nlm.nih.gov/nuccore/MZ379791.1/). The associated Bioproject, Biosample and SRA numbers are PRJNA739476, SAMN19789523, and SRR14868519, respectively.
Additional information
Funding
References
- Aliaga J, Portalatino E, Obregón K, Rodríguez A, Jimenez J. 2019. Presencia del “ají nativo supano” (Capsicum chinense Jacq.) en el valle de Supe, Perú. Peruv Agric Res. 1(2):58–63.
- Arbizu CI, Ellison SL, Senalik D, Simon PW, Spooner DM. 2016. Genotyping-by-sequencing provides the discriminating power to investigate the subspecies of Daucus carota (Apiaceae). BMC Evol Biol. 16(1):1–16.
- Arbizu CI, Simon PW, Martínez-Flores F, Ruess H, Crespo MB, Spooner DM. 2016. Integrated molecular and morphological studies of the Daucus guttatus complex (Apiaceae). Syst Bot. 41(2):479–492.
- Carrizo García C, Barfuss MH, Sehr EM, Barboza GE, Samuel R, Moscone EA, Ehrendorfer F. 2016. Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Ann Bot. 118(1):35–51.
- D’Agostino N, Tamburino R, Cantarella C, De Carluccio V, Sannino L, Cozzolino S, Cardi T, Scotti N. 2018. The complete plastome sequences of eleven capsicum genotypes: insights into DNA variation and molecular evolution. Genes. 9(10):503.
- Doyle JJ, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus. 12(13):39–40.
- Ince AG, Karaca M, Onus AN. 2010. Genetic relationships within and between capsicum species. Biochem Genet. 48(1-2):83–95.
- Jin JJ, Yu WB, Yang JB, Song Y, Depamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1–31.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1):10–12.
- Martínez-Flores F, Arbizu CI, Reitsma K, Juan A, Simon PW, Spooner DM, Crespo MB. 2016. Lectotype designation for seven species names in the Daucus guttatus complex (Apiaceae) from the central and eastern Mediterranean basin. Systematic Botany. 41(2):464–478.
- Meckelmann SW, Riegel DW, van Zonneveld M, Ríos L, Peña K, Ugas R, Quiñonez L, Mueller-Seitz E, Petz M. 2013. Compositional characterization of native Peruvian chili peppers (Capsicum spp.). J Agric Food Chem. 61(10):2530–2537.
- Morales-Soriano E, Kebede B, Ugás R, Grauwet T, Van Loey A, Hendrickx M. 2018. Flavor characterization of native Peruvian chili peppers through integrated aroma fingerprinting and pungency profiling. Food Res Int. 109:250–259.
- Sebastin R, Lee KJ, Cho GT, Shin MJ, Kim SH, Hyun DY, Lee JR. 2019. The complete chloroplast genome sequence of a Bolivian wild chili pepper, Capsicum Eximium Hunz.(Solanaceae). Mitochondr DNA Part B. 4(1):1634–1635.
- Shiragaki K, Yokoi S, Tezuka T. 2020. Phylogenetic analysis and molecular diversity of capsicum based on rDNA-ITS region. Horticulturae. 6(4):87.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Tripodi P, Rabanus-Wallace MT, Barchi L, Kale S, Esposito S, Acquadro A, Schafleitner R, van Zonneveld M, Prohens J, Diez MJ. 2021. Global range expansion history of pepper (Capsicum spp.) revealed by over 10,000 genebank accessions. Proc Natl Acad Sci USA. 118(34):e2104315118.
