Abstract
Holotrichia parallela (Motschulsky, 1854) is an important pest for peanut, potato, and soybean in China, and it causes great economic losses. In this study, we sequenced and analyzed the complete mitochondrial genome (mitogenome) of H. parallela. This mitogenome was 16,975 bp long and encoded 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), and two ribosomal RNA genes (rRNAs). Gene order was conserved and identical to most other previously sequenced Scarabaeidae. Most PCGs of H. parallela have the conventional start codons ATN, with the exception of cox1 (AAC). Except for three genes (cox1, cox2, and cox3) end with the incomplete stop codon T−, all other PCGs terminated with the stop codon TAA or TAG. Phylogenetic analysis positioned H. parallela in a well-supported clade with Rhopaea magnicornis, Polyphylla gracilicornis, and Melolontha hippocastani. The relationships (Dynastinae+(Cetoniinae+(Melolonthinae+(Rutelinae + Scarabaeinae)))) were supported in Scarabaeidae.
The Scarabaeidae is the largest of the 13 families in the Scarabaeoidea and one of the largest and most diverse family of beetles. They are often called scarabs or scarab beetles and consist of over 30,000 species worldwide (Browne and Scholtz Citation2008). The morphological and biological diversity of its members has led to the family being divided into numerous mostly well-defined subfamilies and tribes, as well as into several groups of uncertain taxonomic status (Browne and Scholtz Citation1999). One of its species, the large black chafer Holotrichia parallela (Motschulsky, 1854), is a world-wide pest of many crops, which can cause serious damage to peanut, potato, soybean, and turf grasses, resulting in 15–20% losses annually (Ju et al. Citation2012). Mitogenome can be utilized in research on population genetic structure, taxonomic resolution, phylogeography, and phylogeny. For further study on population genetic structure of H. parallela, we sequenced the complete mitogenome of H. parallela and analyzed the phylogenetic relationships of Scarabaeidae based on mitogenome data.
Male adults of H. parallela were collected from a pig farm of Nanyang City, Henan Province, China (33°06′N, 111°36′E, July 2019) and were stored in Entomological Museum of Henan Institute of Technology (accession number QHU-EHP03, Dr. Jing Zhao, [email protected]). Total genomic DNA was extracted from muscle tissues of the thorax using DNeasy DNA Extraction kit (Qiagen, Hilden, Germany). A pair-end sequence library was constructed and sequenced using Illumina HiSeq 2500 platform (Illumina, San Diego, CA), with 150 bp pair-end sequencing method. A total of 24.6 million reads were generated and had been deposited in the NCBI Sequence Read Archive (SRA) with accession number SRR14149021. Raw reads were assembled using MITObim v 1.7 (Hahn et al. Citation2013). By comparison with the homologous sequences of other Scarabaeidae species from GenBank, the mitogenome of H. parallela was annotated using software GENEIOUS R11 (Biomatters Ltd., Auckland, New Zealand).
The complete mitogenome of H. parallela is 16,975 bp in length (GenBank accession no. MW874410), and contains the typical set of 13 protein-coding, two ribosomal RNA (rRNA) and 22 transfer RNA (tRNA) genes, and one non-coding AT-rich region. Gene order was conserved and identical to most other previously sequenced Scarabaeidae (Cameron et al. Citation2009; Shao et al. Citation2014; Jing et al. Citation2018; Jeong et al. Citation2020; Yang et al. Citation2020). The nucleotide composition of the mitogenome is 75.2% A + T content (A 40.2%, T 35.0%, C 16.2%, and G 8.6%). Four protein-coding genes (PCGs) (nad4, nad4l, nad5, and nad1) were encoded by the minority strand (N-strand) while the other nine were located on the majority strand (J-strand). All PCGs of H. parallela have the conventional start codon for invertebrate mitochondrial PCGs (ATN), with the exception of cox1 (AAC), as the asparagine (AAT or AAC) are proposed to be the start codon for cox1 in suborder Polyphaga (Sheffield et al. Citation2008). Except for three genes (cox1, cox2, and cox3) end with the incomplete stop codon T−, all other PCGs terminated with the stop codon TAA or TAG. The 22 tRNA genes vary from 62 bp (trnC and trnD) to 70 bp (trnQ, trnK, and trnV). Two rRNA genes (rrnL and rrnS) locate at trnL1/trnV and trnV/control region, respectively. The lengths of rrnL and rrnS in H. parallela are 1288 and 755 bp respectively, with AT contents of 78.1% and 72.4%, respectively.
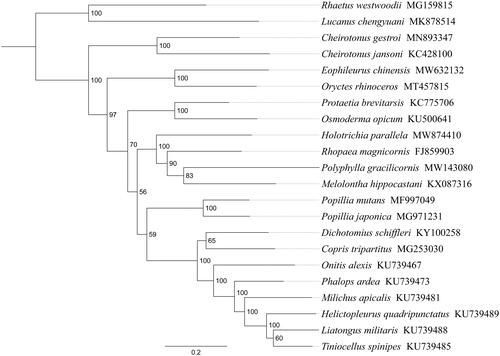
Phylogenetic analysis was performed based on the nucleotide sequences of 13 PCGs from 22 Polyphaga species. Alignments of individual genes were concatenated using SequenceMatrix 1.7.8 (Vaidya et al. Citation2011). Phylogenetic tree was constructed through raxmlGUI 1.5 (Silvestro and Michalak Citation2012). Phylogenetic analysis positioned H. parallela in a well-supported clade with Rhopaea magnicornis, Polyphylla gracilicornis and Melolontha hippocastani (), indicating genus Holotrichia had a close relationship with Rhopaea, Polyphylla, and Melolontha within Melolonthinae. The relationships (Dynastinae+(Cetoniinae+(Melolonthinae+(Rutelinae + Scarabaeinae)))) were supported in Scarabaeidae. The monophyly of Melolonthinae could not be confirmed by this phylogenetic tree. These results provided an important basis for further studies on mitochondrial genome and phylogenetics of Scarabaeidae.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the article.
Data availability statement
The data that support the findings of this study are openly available in NCBI (National Center for Biotechnology Information) at https://www.ncbi.nlm.nih.gov/, reference number MW874410. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA719935, SRR14149021, and SAMN18628722, respectively.
Additional information
Funding
References
- Browne DJ, Scholtz C. 2008. The morphology and terminology of the hindwing articulation and wing base of the Coleoptera, with specific reference to the Scarabaeoidea. Syst Entomol. 19(2):133–143.
- Browne J, Scholtz CH. 1999. A phylogeny of the families of Scarabaeoidea (Coleoptera). Syst Entomol. 24(1):51–84.
- Cameron SL, Sullivan J, Song H, Miller KB, Whiting MF. 2009. A mitochondrial genome phylogeny of the Neuropterida (lace‐wings, alderflies and snakeflies) and their relationship to the other holometabolous insect orders. Zool Scripta. 38(6):575–590.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129.
- Jeong JS, Kim MJ, Kim I. 2020. The mitochondrial genome of the dung beetle, Copris tripartitus, with mitogenomic comparisons within Scarabaeidae (Coleoptera). Int J Biol Macromol. 144:874–891.
- Jing L, Zhou S, Chen Y, Wan X. 2018. Mitogenome of the monotypic genus Rhaetus (Coleoptera: Scarabaeidae: Lucanidae). J Entomol Sci. 53(4):503–513.
- Ju Q, Qu M, Wang Y, Jiang X, Li X, Dong S, Han Z. 2012. Molecular and biochemical characterization of two odorant-binding proteins from dark black chafer, Holotrichia parallela. Genome. 55(7):537–546.
- Shao L, Huang D, Sun X, Hao J, Cheng C, Zhang W, Yang Q. 2014. Complete mitochondrial genome sequence of Cheirotonus jansoni (Coleoptera: Scarabaeidae). Genet Mol Res. 13(1):1047–1058.
- Sheffield N, Song H, Cameron S, Whiting M. 2008. A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol Biol Evol. 25(11):2499–2509.
- Silvestro D, Michalak I. 2012. RaxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12(4):335–337.
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 27(2):171–180.
- Yang C, Zhu E, He Q, Yi C, Hu S, Wang X. 2020. Complete mitochondrial genome of the endangered long-armed scarab Cheirotonus gestroi (Coleoptera: Euchiridae). Mitochondrial DNA B Resour. 5(1):869–870.