Abstract
The first complete chloroplast genome of Campylotropis grandifolia Schindl. is reported and characterized in this study. The whole chloroplast genome was 153,213 bp in length with 128 genes, including 81 protein-coding genes, 39 tRNAs, and eight rRNAs. Maximum-likelihood (ML) phylogenetic analysis of 25 legume species strongly supported that Campylotropis is most closely related with Kummerowia and Lespedeza, which is consistent with previous studies.
Campylotropis Bunge (Fabaceae) is a member of the tribe Desmodieae (Benth.) Hutch. (Iokawa and Ohashi Citation2002a). Among this genus, 38 species are distributed in temperate and tropical regions across Asia, and most species are located in the Southwest China, which is the diversity center of this genus (Lokawa and Ohashi Citation2002a, Citation2002b, Citation2008; Huang et al. Citation2010; Liao and Xu Citation2020). Campylotropis grandifolia Schindl. is known for the quadrangular branches and lack of glandular hairs on the inflorescences (Lokawa and Ohashi Citation2002b). Since few studies have focused on chloroplast genome of Campylotropis, the chloroplast of C. grandifolia can help to resolve the intergeneric and intrageneric relationships in legume species.
The fresh leaves of C. grandifolia were collected from a wasteland in the campus of Southwest Forestry University, Yunnan Province and the voucher specimens were deposited in the herbarium of Ningbo Botanical Garden (NPH, collection #: Kai-Wen Jiang & Wen-Xiang Liu s. n.). Total genomic DNA was extracted with CTAB method, and the genomic library was prepared and paired-end sequenced at Illumina Hiseq 2500 platform (Illumina, San Diego, CA) in Frasergen (Wuhan, China). After filtering the raw data, clean reads were assembled with GetOrganelle v.1.7.0 software (Jin et al. Citation2020) and manually corrected with Geneious software. Annotation was conducted with PGA v. (Qu et al. Citation2019) and OGDRAW (Greiner et al. Citation2019). Finally, the chloroplast genome was deposited in NCBI with the accession of MZ918589.
The total length of chloroplast genome of C. grandifolia was 153,213 bp, the total GC content is 35% the specific length and the GC content of each part is shown in . A total of 111 species and 128 genes were annotated in C. grandifolia as shown in .
Table 1. Base composition of chloroplast genome in Campylotropis grandifolia.
Table 2. Genes present in chloroplast genome of Campylotropis grandifolia.
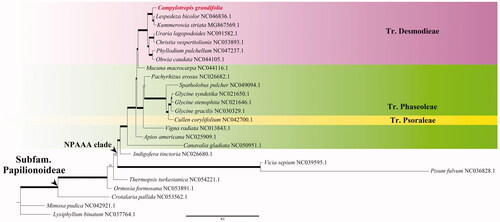
In order to confirm the systematical position of C. grandifolia, a total of 24 legume chloroplast genomes were downloaded from GenBank and applied to construct the phylogenetic tree, where Lysiphyllum binatum (Blanco) de Wit was selected as the outgroup. Specially, we aligned these 25 cp genomes with MAFFT v.7.481 (Katoh and Standley Citation2013), eliminated the gaps with MEGA X (Kumar et al. Citation2018), and constructed maximum-likelihood (ML) tree with IQ-TREE v.1.6.12 (Nguyen et al. Citation2015) in K3Pu + F+R5 model (). Our results showed that C. grandifolia is sister to the clade consisting of Kummerowia striata (Thunb.) Schindl. and Lespedeza bicolor Turcz., which is congruent with previous study using the combined cpDNA fragments data (Xu et al. Citation2012).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in The DNA Data Bank of Japan (DDBJ; http://www.ddbj.nig.ac.jp) under the accession no. MZ918589. The associated BioProject and Bio-Sample numbers are PRJNA743046 and SAMN20688300, respectively.
Additional information
Funding
References
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Huang PH, Ohashi H, Iokawa Y. 2010. Camyplotropis. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China 10. Beijing and St. Louis: Science Press and Missouri Botanical Garden Press; p. 292–302.
- Iokawa Y, Ohashi H. 2002a. A taxonomic study of Campylotropis (Leguminosae) I. J Jpn Bot. 77(4):179–222.
- Iokawa Y, Ohashi H. 2002b. A taxonomic study of Campylotropis (Leguminosae) II. J Jpn Bot. 77(5):251–283.
- Iokawa Y, Ohashi H. 2008. Campylotropis (Leguminosae) of China, an enumeration and distribution. J Jpn Bot. 83(1):36–59.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 35(6):1547–1549.
- Liao M, Xu B. 2020. Campylotropis albopubescens stat. nov. (Leguminosae: Papilionoideae: Desmodieae): the only species in the genus reproduced via rootstocks. Phytotaxa. 454(3):226–230.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Qu X-J, Moore MJ, Li D-Z, Yi T-S. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Xu B, Wu M, Gao X-F, Zhang L-B. 2012. Analysis of DNA sequences of six chloroplast and nuclear genes suggests incongruence, introgression, and incomplete lineage sorting in the evolution of Lespedeza (Fabaceae). Mol Phylogenet Evol. 62(1):346–358.