Abstract
In this study, we presented the complete mitochondrial genome of a tropical midge Chironomus kiiensis Tokunaga by Illumina sequencing technology. The complete mitogenome of C. kiiensis is 15,710 bp in length, including 13 protein-coding genes (PCGs), two ribosomal RNAs and 22 tRNAs. The overall nucleotide composition is A: 39.3%, T: 37.6%, C: 13.6%, and G: 9.5%. Most PCGs use ATN as the start codon excluding COX1 (TTG), and most PCGs use TAA as the stop codon excluding ND4 (TAG). Gene order of the C. kiiensis mitogenome is identical to those of other Chironomidae mitogenomes. Phylogenetic analysis based on the mitogenomic PCGs indicate that Chironomus is a sister clade to Polypedilum.
Chironomus species, from the dipteran family Chironomidae, are widely distributed in the northern hemisphere at temperate latitudes (Péry et al. Citation2005). The tropical midge Chironomus kiiensis larvae are usually found in organically enriched waters and broadly employed in aquatic toxicity studies (Sun et al. Citation2019), whereas their mitochondrial genomes have not been described. Following the reports of C. yoshimatsui and C. flaviplumus mitochondrial genomes (Hiki et al. Citation2021; Park et al. Citation2021), this study described the complete mitochondrial genome of C. kiiensis, to contribute to further study of Chironomus.
The samples of C. kiiensis were collected from the sediments of a pond at Lishui, Zhejiang, China (28°39′30′′N, 120°5′29′′E) on August 2019. The specimens are deposited at the College of Life Sciences, Taizhou University (www.tzc.edu.cn, contact person: Xin Qi; e-mail: [email protected]) under the voucher number BSZ21. Genomic DNA was extracted from a larva abdomen using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) and next-generation sequencing was performed at Illumina NovaSeq 6000 platform (Illumina, San Diego, CA). Raw reads were filtered by Trimmomatic version 0.39 (Bolger et al. Citation2014) and assembled by SPAdes version 3.14.1 (Bankevich et al. Citation2012). The assembly mitochondrial genome sequence was annotated with MITOS web server (Bernt et al. Citation2013) and tRNAscan-SE (Lowe and Chan Citation2016).
The complete mitochondrial genome of C. kiiensis is 15,710 bp in length, including 13 protein-coding genes (PCGs), 2 ribosomal RNAs and 22 tRNAs. The overall nucleotide composition is A: 39.3%, T: 37.6%, C: 13.6%, and G: 9.5%. Most PCGs use ATN as the start codon excluding COX1 (TTG), and most PCGs use TAA as the stop codon excluding ND4 (TAG). Gene arrangement of the C. kiiensis mitogenome is identical to those of other known Chironomidae mitogenomes.
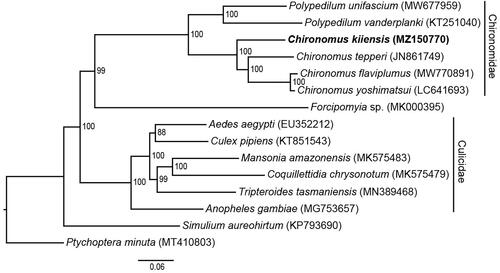
To discuss the phylogenetic position of C. kiiensis, 14 mitochondrial genomes were downloaded from NCBI, including 13 Culicomorpha species as ingroups and one Ptychopteromorpha species Ptychoptera minuta as an outgroup. The PCGs sequences were multiple aligned using MAFFT method (Katoh Citation2002), and conserved blocks were identified by Gblocks version 0.91b (Talavera and Castresana Citation2007). The phylogenetic relationship was reconstructed using maximum-likelihood (ML) method under GTR + I + G substitution model for 1000 bootstraps via RAxML version VI (Stamatakis Citation2014). The resulting tree showed that three Chironomus species were in a cluster and the genus was closely related to Polypedilum ().
Figure 1. Maximum-likelihood (ML) phylogeny of 14 registered Culicomorpha mitogenomes including C. kiiensis, and Ptychoptera minuta as an outgroup based on the concatenated sequences of mitogenomic PCGs. The phylogenetic analysis was performed under GTR + I + G substitution model for 1000 bootstraps. The numbers at the nodes indicate the ML bootstrap values.
Disclosure statement
The authors report no conflicts of interest and are alone responsible for the content and writing of this paper.
Data availability statement
The data support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov. The complete mitochondrial genome of Chironomus kiiensis for this study has been deposited in GenBank with accession number MZ150770. The associated BioProject, BioSample and SRA numbers are PRJNA705522, SAMN22359574, and SRR16381213, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Hiki K, Oka K, Nakajima N, Yamamoto H, Yamagishi T, Sugaya Y. 2021. The complete mitochondrial genome of the non-biting midge Chironomus yoshimatsui (Diptera: Chironomidae). Mitochondrial DNA B Resour. 6(10):2995–2996.
- Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Park K, Kim W-S, Park J-W, Kwak I-S. 2021. Complete mitochondrial genome of Chironomus flaviplumus (Diptera: Chironomidae) collected in Korea. Mitochondrial DNA B Resour. 6(10):2843–2844.
- Péry ARR, Mons R, Garric J. 2005. Modelling of the life cycle of Chironomus species using an energy-based model. Chemosphere. 59(2):247–253.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Sun L, Wang J, Li X, Cao C. 2019. Effects of phenol on glutathione S-transferase expression and enzyme activity in Chironomus kiiensis larvae. Ecotoxicology. 28(7):754–762.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577.
