Abstract
The freshwater gastropod Tarebia granifera (Lamarck, 1816) is found in Taiwan, Hainan, and Guangdong provinces in China, and is one of the main intermediate hosts of trematodes that infect humans. The taxonomic positions of some cerithioidean families are still unclear, and whole mitochondrial genome studies are scarce in the Thiaridae. In this study, we describe the complete mitogenome of Tarebia granifera (Lamarck, 1816). The mitogenome is 15,555 bp in length, with a total of 37 genes, including 13 protein-coding genes, 2 rRNA genes, and 22 tRNA genes. It is consistent with the essential features of previously studied mitochondrial genomes of species belonging to the superfamily Cerithioidea. Our study demonstrates the usefulness of mitogenomic data for resolving phylogenetic relationships of families within Cerithioidea and may also contribute to the prevention and control of the parasitic diseases caused by trematodes, which use T. granifera as an intermediate host.
The superfamily Cerithioidea currently includes 21 families (Bouchet et al. Citation2017; Neiber and Glaubrecht Citation2019d), nine of which inhabit freshwater environments: Amphimelaniidae, Hemisinidae, Melanopsidae, Pachychilidae, Paludomidae, Pleuroceridae, Semisulcospiridae, Thiaridae, and Zemelanopsidae (Campbell Citation2019; Glaubrecht and Neiber Citation2019a, Citation2019b; Neiber and Glaubrecht Citation2019a, Citation2019b, Citation2019c, Citation2019d; Strong and Lydeard Citation2019). Initially, most of the freshwater species in the superfamily Cerithioidea were placed in the family Melaniidae and the genus Melania (Neiber and Glaubrecht Citation2019b). With the development of molecular systematics, it was found that the cerithioidean freshwater taxa were not monophyletic, and the taxonomic positions of some cerithioidean families are still unclear (Strong et al. Citation2011; Neiber and Glaubrecht Citation2019d). Tarebia granifera (Lamarck, 1816) is an invasive species native to South and Southeast Asia and several Western Pacific Islands. In addition, the species has become widely invasive in the tropics outside its native range (e.g. Africa, the Mediterranean region and the Middle East, as well as North, Central and South America), with the spreading being attributed to the aquarium trade or dispersal by birds (Malatji et al. Citation2021; Chalkowski et al. Citation2021). In China, T. granifera is mainly distributed in the provinces of Taiwan, Hainan, and Guangdong (Liu et al. Citation1993). Tarebia granifera is one of the main intermediate hosts of Paragonimus westermani and Metagomimus yokogawai (Liu et al. Citation1993; Veeravechsukij et al. Citation2018a, Citation2018b). The complete mitochondrial genome of this species is still lacking. By sequencing the mitochondrial genome of T. granifera, our study provides additional information on mitogenome evolution and the phylogeny of Cerithioidea, and may also contribute to the prevention and control of the parasitic disease caused by trematodes, which use T. granifera as an intermediate host.
Specimens of Tarebia granifera were collected from Pingshan Wetland Park (22°41′53″N, 114°22′4″E), Shenzhen, China, in 2019. Morphological identification followed the literature (Liu et al. Citation1993) and specimens from the Institute of Zoology, Chinese Academy of Sciences. Tissues were preserved at −80 °C in a refrigerator, and the voucher specimen (number: 21-NCU-XPWU-TG01; contact Xiao-Ping Wu: [email protected]) was deposited in the Museum of Biology in Nanchang University. Referring to the previous study, the mitogenome of Tarebia granifera was sequenced using primer-walking (Xie et al. Citation2019). Firstly, cob (Merritt et al. Citation1998), rrnL (Palumbi et al. Citation1991), and cox1 (Folmer et al. Citation1994) genes were obtained by universal primers. Secondly, three sets of primers were designed to amplify the complete mitogenome into three long fragments. Details of primers and PCR conditions are shown in Supporting information S1. Finally, sequences were assembled into the whole mitogenome by SeqMan program (DNAstar).
ARWEN (Laslett and Canbäck Citation2008) and MITOS (Bernt et al. Citation2013) were used jointly to identify localized transfer RNA (tRNA) genes and analyze secondary structures. Ribosomal RNAs (12S rRNA and 16S rRNA) were then compared by homology with other Cerithioidea species. Open Reading Frame Finder (ORF Finder) (http://www.ncbi.nlm.nih.gov/orffinder/) and BLAST searches were used to identify 13 protein-coding genes. We downloaded 12 published gastropod mitogenomes from GenBank, and used Tricula hortensis (Truncatelloidea: Pomatiopsidae) as outgroup. Phylogenetic analysis was conducted for Bayesian inference (BI) in MrBayes (Ronquist et al. Citation2012) based on the best-fit partitioning schemes and models (details in Table S2) selected by PartitionFinder (Lanfear et al. Citation2017). Two simultaneous runs with four independent chains were implemented for 10 million generations, sampling every 1000 generations. The first 25% of these trees were discarded as burnin.
The mitogenome of Tarebia granifera is 15,555 bp in length (GenBank accession number: MZ662113). It contains 37 genes that encode two rRNAs, 22 tRNAs, and 13 proteins. The mitogenome base composition was A (30.9%), T (34.5%), C (17.7%), and G (16.9%), with the A+T content of 65.4% being remarkably higher than the G+C content (34.6%), which was similar to other Cerithioidea (Lee et al. Citation2019). The total length of protein-coding genes is 11,226 bp, accounting for 72.17% of the complete mitochondrial genome. The 12S rRNA (958 bp) gene was located between the trnS and trnT genes and the 16S rRNA (1,354 bp) gene was located between the trnV and trnL genes, respectively. This mitogenome contained 22 tRNA genes, with the shortest tRNA genes trnY, trnH and trnM being 65 bp in length, and the longest tRNA genes trnW, trnA and trnE being 70 bp in length.
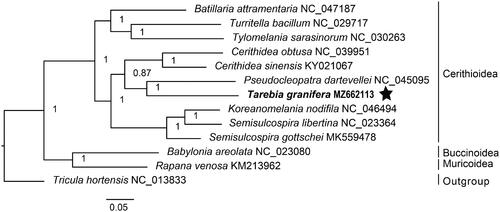
The tree topology obtained in this study is consistent with previous studies (Wang et al. Citation2017; Lee et al. Citation2019). Phylogenetic analysis based on 12 PCGs and two rRNA genes showed that Tarebia granifera and Pseudocleopatra dartevellei had a close relationship (Bayesian posterior probability = 1), forming the sister cluster of Cerithidea sinensis and Cerithidea obtusa (). The majority of the tree nodes were well-resolved with high posterior probabilities. Our results demonstrate the usefulness of complete mitochondrial genome in resolving phylogenetic relationships among cerithioidean lineages.
Figure 1. Bayesian 50% majority-rule consensus tree of 10 cerithioidean gastropods based on 12 mitochondrial PCGs and two rRNA genes. The analysis included also representatives of the superfamilies Buccinoidea and Muricoidea. Tricula hortensis (Truncatelloidea) was used as outgroup.
Additional supporting information may be found online in the Supporting information section at the end of the article.
Ethical approval
The handling of freshwater snails was conducted in accordance with the guidelines on the care and use of animals for scientific purposes set by the Institutional Animal Care and Use Committee (IACUC) of Nanchang University, Jiangxi, China.
Author contributions
N.Y. and X.C.H. designed the study. N.Y. and S.Z. performed the laboratory work. N.Y. analyzed the data, prepared figures and tables, and wrote the paper. X.C.H. supervised the molecular analyses. S.Oy. and X.P.W. revised the manuscript. All authors contributed to the critical review and revision of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ662113.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bouchet P, Rocroi JP, Hausdorf B, Kaim A, Kano Y, Nützel A, Parkhaev P, Schrödl M, Strong EE. 2017. Revised classification, nomenclator and typification of gastropod and monoplacophoran families. Malacologia. 61(1-2):1–527.
- Campbell DC. 2019. Semisulcospiridae Morrison, 1952. In: Lydeard C, Cummings KS. (Eds.), Freshwater Mollusks of the World – A Distribution Atlas. Baltimore: Johns Hopkins University Press; p. 81–85.
- Chalkowski K, Morgan A, Lepczyk CA, Zohdy S. 2021. Spread of an Avian Eye Fluke, Philophthalmus gralli, through Biological Invasion of an Intermediate Host. J Parasitol. 107(2):336–348.
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 3(5):294–299.
- Glaubrecht M, Neiber MT. 2019a. Hemisinidae Fischer and Crosse, 1891. In: Lydeard C, Cummings KS. (Eds.), Freshwater Mollusks of the World – A Distribution Atlas. Baltimore: Johns Hopkins University Press; p. 51–55.
- Glaubrecht M, Neiber MT. 2019b. Thiaridae Gill, 1873 (1823). In: Lydeard C, Cummings KS. (Eds.), Freshwater Mollusks of the World – A Distribution Atlas. Baltimore: Johns Hopkins University Press; p. 86–89.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Lee SY, Lee HJ, Kim YK. 2019. Comparative analysis of complete mitochondrial genomes with Cerithioidea and molecular phylogeny of the freshwater snail, Semisulcospira gottschei (Caenogastropoda, Cerithioidea). Int J BiolMacromol. 135:1193–1201.
- Liu YY, Zhang WZ, Wang YX. 1993. Medical malacology. Beijing: Ocean Press.
- Malatji MP, Myende N, Mukaratirwa S. 2021. Are Freshwater Snails, Melanoides sp. and Invasive Tarebia granifera (Gastropoda: Thiaridae) Suitable Intermediate Hosts for Calicophoron microbothrium (Trematoda: Paramphistomoidea)? An Experimental Study. Front Vet Sci. 8:705954.
- Merritt TJ, Shi L, Chase MC, Rex MA, Etter RJ, Quattro JM. 1998. Universal cytochrome b primers facilitate intraspecific studies in molluscan taxa. Mol Mar Biol Biotechnol. 7(1):7–11.
- Neiber MT, Glaubrecht M. 2019b. Pachychilidae Fischer and Crosse, 1892. In: Lydeard C, Cummings KS. (Eds.), Freshwater Mollusks of the World – A Distribution Atlas. Baltimore: Johns Hopkins University Press; p. 62–67.
- Neiber MT, Glaubrecht M. 2019c. Paludomidae Stoliczka, 1868. In: Lydeard C, Cummings KS. (Eds.), Freshwater Mollusks of the World – A Distribution Atlas. Baltimore: Johns Hopkins University Press; p. 68–73.
- Neiber MT, Glaubrecht M. 2019d. Unparalleled disjunction or unexpected relationships? Molecular phylogeny and biogeography of Melanopsidae (Caenogastropoda: Cerithioidea), with the description of a new family and a new genus from the ancient continent Zealandia. Cladistics. 35(4):401–425.
- Neiber MT, Glaubrecht M, 2019a. Melanopsidae H. Adams and A. Adams, 1854. In: Lydeard C, Cummings KS. (Eds.), Freshwater Mollusks of the World – A Distribution Atlas. Baltimore: Johns Hopkins University Press; p. 56–61.
- Palumbi SR, Martin A, Romano S, McMillian WO, Stice L, Grabowski G. 1991. The simple fool’s guide to PCR. Honolulu: University of Hawaii.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Strong EE, Colgan DJ, Healy JM, Lydeard C, Ponder WF, Glaubrecht M. 2011. Phylogeny of the gastropod superfamily Cerithioidea using morphology and molecules. Zool J Linn Soc. 162(1):43–89.
- Strong EE, Lydeard C. 2019. Pleuroceridae P, Fischer, 1885. In: Lydeard C, Cummings KS. (Eds.), Freshwater Mollusks of the World – A Distribution Atlas. Baltimore: Johns Hopkins University Press; p. 74–80.
- Veeravechsukij N, Krailas D, Namchote S, Wiggering B, Neiber MT, Glaubrecht M. 2018a. Molecular phylogeography and reproductive biology of the freshwater snail Tarebia granifera in Thailand and Timor (Cerithioidea, Thiaridae): morphological disparity versus genetic diversity. Zoosyst. Evol. 94(2):461–493.
- Veeravechsukij N, Namchote S, Neiber MT, Glaubrecht M, Krailas D. 2018b. Exploring the evolutionary potential of parasites: larval stages of pathogen digenic trematodes in their thiarid snail host Tarebia granifera in Thailand. Zoosyst. Evol. 94(2):425–460.
- Wang JG, Zhang D, Jakovlic I, Wang WM. 2017. Sequencing of the complete mitochondrial genomes of eight freshwater snail species exposes pervasive paraphyly within the Viviparidae family (Caenogastropoda). PLOS One. 12(7):e181699.
- Xie GL, Köhler F, Huang XC, Wu RW, Zhou CH, Ouyang S, Wu XP. 2019. A novel gene arrangement among the Stylommatophora by the complete mitochondrial genome of the terrestrial slug Meghimatium bilineatum (Gastropoda, Arionoidea). Mol Phylogenet Evol. 135:177–184.
