Abstract
Rhaponticum uniflorum is commonly used as a source for traditional medicines with the main effect of clearing heat. Here, we sequenced the complete chloroplast (cp) genome of R. uniflorum to develop molecular markers for taxonomic classification and species determination of R. uniflorum. It was 152,760 bp in size and has a typical circular structure, including a pair of inverted repeats with 25,205 bp, a large single-copy region with 83,687 bp, and a small single copy region with 18,663 bp. The genome encodes 110 unique genes, including 80 protein-coding, four rRNA and 26 tRNA genes. Phylogenomic analysis shows that R. uniflorum is closely related to the Saussurea. The study is useful for phylogenetic and population genetic studies of Rhaponticum plants.
Rhaponticum uniflorum (L.) DC. (Rhaponticum uniflorum (L.) DC. 1810) is a perennial herb and belongs to the Rhaponticum genus, Asteraceae family. The root, used directly as a traditional medicine, has the effect of clearing heat, detoxifying, relieving swelling and expelling pus (Committee Citation2020). Rhaponticum uniflorum contains several types of compounds including phytoecdysones, steroids, terpenoids, thiophenes, and flavones (Zhu et al. Citation1991). Pharmacological studies showed its activities of anti-inflammatory, anti-oxidative, immunomodulating, and anti-tumor effects (Jin et al. Citation2011; Chen et al. Citation2017). However, there is little research on the genomic resource of R. uniflorum, which limited the species identification and resource conservation. In this study, we report the cp-genome sequence of R. uniflorum, which is the first report of chloroplast genome in Rhaponticum.
A sample was collected in Dafutou village (Geographic coordinates 115°95′19″N, 40°38′42″E), Zhangshanying Town, Yanqing District, Beijing, China and identified as R. uniflorum by Professor Jinwen You. A specimen and its DNA were deposited at Institute of Medicinal Plant Development (www.implad.ac.cn/, Chang Liu and [email protected]) under the voucher number 2019071712. Genomic DNA was extracted from mature fresh leaves through modified CTAB method (Doyle Citation1987), and purified to construct a DNA library with the insertion size of 500 bp. Paired-end sequencing was performed on the Hiseq 2500 platform (Illumina, San Diego, CA, USA). The raw sequence data were fed into NOVOPlasty (version 2.7.2) (Dierckxsens et al. Citation2017) to assemble the cp-genome. Then, the annotation was finished by using CpGAVAS2 (Shi et al. Citation2019).
The chloroplast genome of R. uniflorum (accession number MW683229) is 152,760 bp in size with a pair of inverted repeats (IRs) of 25,205 bp separated by a large single-copy (LSC) region of 83,687 bp and a small single-copy (SSC) region of 18,663 bp. The chloroplast genome encoded129 genes, of which 110 are unique genes including 80 protein-coding, four ribosome RNA (rRNA), and 26 transfer RNA (tRNA) genes. Among them, ten protein-coding genes had one intron and two had two introns. Six tRNA genes were found to contain one intron. The GC content of the genome was 51.47%, of which the protein-coding, the rRNA, and the tRNA genes were 70.6, 55.09, and 51.78%, respectively.
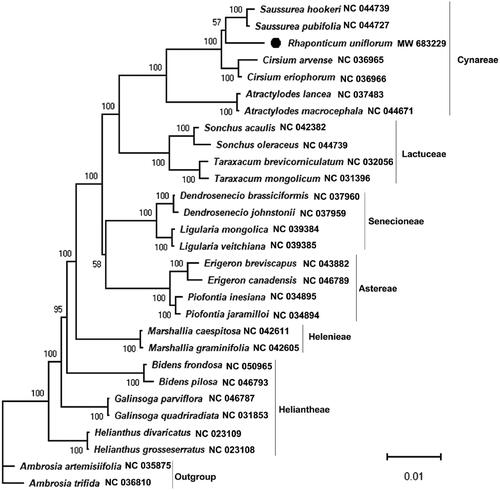
To infer the phylogenetic position of R. uniflorum, we constructed a maximum-likelihood (ML) tree using the sequences of the whole chloroplast genomes of twenty-nine species in the Asteraceae family, including two species in the Ambrosia used as outgroup. In the phylogenetic tree, R. uniflorum is the only species in the Rhaponticum, and each of the other genera has two species. 44 shared proteins sequences were subjected to PhyloSuite to construct the ML phylogenomic tree (Zhang et al. Citation2020). Then, the phylogenomic tree was visualized by using MEGA X (Kumar et al. Citation2018). As shown in , R. uniflorum is clustered within the group consisting Saussurea hookeri and S. pubifolia, and the bootstrap score is greater than 50 and less than 70, indicating that R. uniflorum is very closely related to Saussurea. Except R. uniflorum, all two species from the same genus are clustered together, and some species from the same tribe are clustered together. This study will be useful in phylogenetic and population genetic studies of Rhaponticum plants.
Figure 1. Molecular phylogenetic tree showing the position of Rhaponticum uniflorum in the family Asteraceae based on the complete chloroplast genomes among 29 species. The tree was constructed using maximum likelihood (ML) algorithm. Numerical value beside each node shows the bootstrap value obtained from 1000 replications. The branch lengths are scaled with a scale bar. The GenBank accession number for the corresponding sequences is shown to the right of the Latin name.
Ethics approval and consent to participate
The research, including the collection of plant materials, was carried out in accordance with guidelines provided by the authors’ institutions and national or international regulations.
Authors’ contributions
The article was designed and conceived by Chang Liu and Chengxi Wei; It was Liqiang Wang, HuBoqin, Qiang Zhou, Mei Jiang and Guohua Gong who got involved in the analysis and interpretation of the data; HuBoqin and Qiang Zhou drafted the article; Chang Liu revised it critically for intellectual content; All authors approved the final version to be published and agreed to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession NO. MW683229. The associated BioProject, BioSample, and SRA number are PRJNA722731 and SAMN18790581, and SRR14270266 respectively.
Additional information
Funding
References
- Chen H, Wang C, Qi M, Ge L, Tian Z, Li J, Zhang M, Wang M, Huang L, Tang X. 2017. Anti-tumor effect of Rhaponticum uniflorum ethyl acetate extract by regulation of peroxiredoxin1 and epithelial-to-mesenchymal transition in oral cancer. Front Pharmacol. 8:870.
- Committee NP. 2020. Pharmacopoeia of the People’s Republic of China (2020). Vol. 2. Beijing: Chemistry Industry Press; p. 387–388.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19:11–15.
- Jin AH, Hui-Xian XU, Liu WJ, Quan JS, Yin XZ. 2011. Studies on anti-tumor effect and mechanism of Rhaponticum uniflorum in H22-bearing mice. Chinese J Exp Tradit Med Form. 17(5):165–167.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
- Zhu L, Lu Y, Chen D. 1991. Composition of essential oil from inflorescences of Rhaponticum uniflorum (L.) DC. Zhongguo Zhong Yao Za Zhi. 16(12):739–740, 762–763.
