Abstract
Danio roseus, collected from Gadu River in Yingjiang area, Yunnan Province, China, had been sequenced on Illumina HiSeq platform with 16523 bp in length, which included 37 genes encoding 13 proteins-coding genes (PCGs), 22 tRNAs, 2 rRNAs and a displacement loop region. The proportion of nucleotides in mitochondrial genome was T (28.7%), C (23.2%), A (32.3%), G (15.8%) with an AT bias of 61%. Maximum-Likelihood phylogenetic tree was reconstructed using concatenated mitochondrial protein-coding genes of D. roseus and other 12 fishes. The result of phylogenetic analysis supported the consanguineous relationship among D. roseus, D. rerio, D. nigrofasciatus and D. dangila.
The Danio roseus (Fang and Kottelat Citation2000) belongs to the subfamily Danioninae, family Danionidae, which is distributed in Yunnan Province of China; Lao People's Democratic Republic; Myanmar and Thailand. The characteristic description of the D. roseus is similar to that of D. albolineatus (Chu & Chen Citation1989). This project is the first to determine the mitochondrial genome sequence of D. roseus, which would be helpful to the study of its phylogenetic relationship and species identification.
The samples were obtained from Gadu River in Yingjiang area, Yunnan Province, China (24°60′81.86″ N, 97°66′19.38″E) on Jun 5th and stored at Aquatic Science and Technology Institution Herbarium (voucher ASTIH-21b0616d01, https://www.jsahvc.edu.cn/, Person in charge: Xiao Jiang CHEN, email: [email protected]). Fresh samples were stored in dry ice and transported to Shanghai Genesky Biotechnologies Inc. The total genomic DNA was extracted from muscle tissues using Tguide Cell/tissue genomic DNA Extraction Kit (OSR-M401) (Tiangen, Beijing, China). The Illumina HiSeq 4000 Sequencing platform (Illumina, CA, USA) was used to perform the mitochondrial genome sequence. We used the software MetaSPAdes (Nurk et al. Citation2017) to assemble the sequence and annotated the genome with the MitoMaker software (Bernt et al. Citation2013). Phylogenetic tree was based on MEGA-X software (Tamura et al. Citation2013). The animal experimentswere approved by the Ethical Committee for Animal Experiments of Jiangsu Agri-animal Husbandry Vocational College and conducted in accordance with the Guidelines for Experimental Animals of the Ministry of Science and Technology (Beijing, China).
The complete mitochondrial genome of D. roseus was 16523 bp in length, which contained 37 genes encoding 13 proteins, 22 tRNAs, 2 rRNAs and 1 displacement loop region (D-loop) located between tRNAPhe and tRNAPro with 913 bp in size. The proportion of nucleotides in mitochondrial genome was T (28.7%), C (23.2%), A (32.3%), G (15.8%) with an AT bias of 61%. Among all 37 genes, 28 genes were encoded on the H-strand while tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer(UCN), tRNAGlu, tRNAPro and ND6 were encoded on L-strand. The characteristics of gene overlap and segregation were also observed in the mitochondrial genome of D. roseus. The interval length of 17 genes was 100 bp, ranging from 1 to 32 bp, and the overlap length of 9 genes was 28 bp, with overlapping base numbers between 1 and 7 bp ().
Table 1. Relevant features of Danio roseus mitochondrial genome.
Among 13 protein-coding genes (PCGs), the longest gene was ND5 (1800 bp) and the shortest gene was ATP8 (165 bp). Except that the COI gene took GTG as starting codon, the other 12 genes took ATG as starting codon. The PCGs (ND1, COI, Atp8, Atp6, COIII, ND5, and ND4L) took TAA as termination codon while two genes (CO II, Cytb) ended with incomplete termination codon (T) respectively, nevertheless the genes (ND3, ND6) ended with complete termination codon (TAG, AGG) respectively. The length of tRNA ranged from 68 to 75 bp. The 12S rRNA was 954 bp in length as well as 1643 bp of the 16S rRNA.
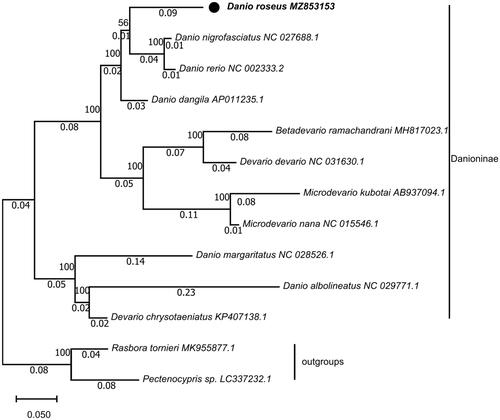
The sequence had been submitted to NCBI (NO. MZ853153). To find out the phylogenetic position of D. roseus, the mitochondrial genome sequences of 12 reported fishes were downloaded from NCBI GenBank, and 13 protein-coding genes (PCGs) of each fish were concatenated, respectively. Evolutionary analyses were conducted in MEGA X (Kumar et al. Citation2018) and the evolutionary history was inferred by using the Maximum Likelihood method and General Reversible Mitochondrial + Freq. model (Adachi and Hasegawa Citation1996). Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT + G + F model, with a bootstrap of 1000 replicates. The result of phylogenetic analysis showed D. rerio and D. nigrofasciatus grouped together, and they formed a sister-group with D. roseus, then the above three formed a sister-group with D. dangila. The analysis of phylogenetic tree supported the consanguineous relationship among D. roseus, D. rerio, D. nigrofasciatus, and D. dangila (). Research on phylogeny of Danio suggested that there was a very close relationship among D. albolineatus, D. rerio, and D. nigrofasciatus (Fang Citation2003). The complete mitochondrial genome of D. roseus would be beneficial for resource conservation and phylogeny.
Figure 1. Maximum-likelihood (ML) phylogenetic tree reconstructed using concatenated mitochondrial protein-coding genes of D. roseus and other 12 fishes. Pectenocypris sp. and Rasbora tornieri were used as outgroups. Accession numbers are indicated after the species names. The tree topology was evaluated by 1000 bootstrap replicates. Bootstrap values at the nodes correspond to the support values for ML methods.
Author contributions
Lin Song and Xiao Jiang Chen make substantial contributions to the conception or design of the work, and drafting the paper, and final approval of the version to be published and agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved; Xing Peng Han and Yi Wen Gu were involved in the acquisition, analysis and interpretation of the data; the drafting of the paper, and the final approval of the version to be published, and agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The contributions are ranked in order.
Acknowledgment
The authors thank Dr. Li Yanping of Neijiang Normal University for her help and acknowledge the Shanghai Genesky Biotechnologies Inc., for their technical support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ853153
The associated "BioProject", "Bio-Sample" and "SRA" numbers are PRJNA769947, SAMN22183819, SRR16277915, respectively.
Additional information
Funding
References
- Adachi J, Hasegawa M. 1996. Model of amino acid substitution in proteins encoded by mitochondrial DNA. J Mol Evol. 42(4):459–468.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chu XL, Chen YR. 1989. Ichthyology of Yunnan, China. Beijing, China: Science Press.
- Fang F, Kottelat M. 2000. Danio roseus, a new species from the Mekong basin in northeastern Thailand and northwestern Laos (Teleostei, Cyprinidae). Ichthyol Explor Fres. 11(2):149–154.
- Fang F. 2003. Phylogenetic analysis of the Asian Cyprinid genus Danio (Teleostei, Cyprinidae). Copeia. 2003(4):714–728.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. MetaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
