Abstract
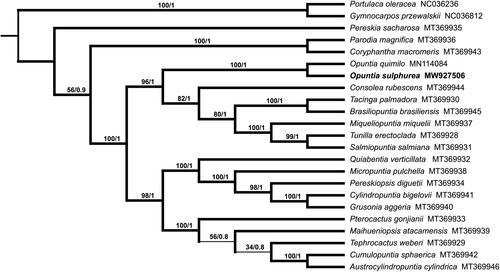
Opuntia sulphurea Gillies ex Salm-Dyck 1834 (Cactaceae) acts as an invasive species due to its ability to survive in various environments. In this study, we assembled the complete chloroplast (cp) genome of Opuntia sulphurea, which was 122,740 bp in length. The genome contained 100 genes, including 65 protein-coding genes, 31 tRNA genes and four rRNA genes. The base composition of the chloroplast genome was 32.11% A, 17.74% G, 18.34% C, and 31.80% T, resulting in an overall G + C content of 35.39%. A phylogenetic analysis across 23 species in Caryophyllales demonstrated a close relationship between Opuntia sulphurea and Opuntia quimilo.
Opuntia sulphurea Gillies ex Salm-Dyck 1834, which belongs to the cacti family Cactaceae, is an endemic species mainly distributed in the arid region of Argentina (Carreira et al. Citation2014). Commonly known as ‘prickly pear’ cacti, O. sulphurea has been widely used as food, forage, and medicine by natives in the western hemisphere for thousands of years (Pimienta-Barrios Citation1994; Casas and Barbera Citation2002; Panico et al. Citation2007). Thus, the genomic information is urgently needed to help defend the invasion of O. sulphurea.
Succulent stems were collected from an individual of O. sulphurea, which was cultivated in Minqin Desert Botanical Garden, Wuwei City, China (38°35′6.95″N, 102°58′22.68″E); Collector: Shengdan Wu ([email protected]). The specimen is deposited in the herbarium of Lanzhou University, Lanzhou, China under the voucher number MQXRZ01 (person in charge: Shengdan Wu: [email protected]). Genomic DNA was isolated using the modified CTAB method (Allen et al. Citation2006). The 20.45 Gb whole-genomic sequences were obtained using the Illumina HiSeq 2500 platform (Illumina, San Diego, CA). Chloroplast genome assembly was performed by using the NOVOPlasty software (Dierckxsens et al. Citation2016). Annotation was conducted with Plastid Genome Annotator (PGA, Qu et al. Citation2019).
The complete chloroplast genome of O. sulphurea (GenBank: MW927506) was 122,740 bp in length, and we found there are no inverted repeats (IRs) existed, which is differ from the chloroplast genome of O. quimilo (Köhler et al. Citation2020). It contains 100 genes, including 65 protein-coding genes, four rRNA genes, and 31 tRNA genes. Four rRNA genes are rrn5, rrn4.5, rrn23, and rrn16.
We also constructed the phylogenetic trees with the maximum likelihood (ML, RaxML version 8) (Stamatakis Citation2014), and Bayesian analysis (BI, MrBayes version 3.2) (Ronquist et al. Citation2012) methods. The alignments were created by the MAFFT (Katoh and Standley Citation2013) using 23 related species of Caryophyllales including the Gymnocarpos przewalskii and Portulaca oleracea as outgroup (). The results indicated that all 21 species from the Cactaceae were clustered together and O. sulphurea was most closely related to O. quimilo. This complete chloroplast genome can be readily used for population genomic studies of O. sulphurea, and such information would serve as fundamental for further research on defense to its invasion into other ecosystems.
Author contributions
S.W. collected samples; J.C, S.Z., X.D., W.T and Y.Y. conducted the analyses and interpreted the results; J.C., S.Z. and S.W. wrote the manuscript; all authers read and agreed the final version of the manuscript; S.W. designed the study.
Sampling permission statement
The plant is not listed in the List of State Key Protected Wild Plants of P.R. China (No.5 2021, by National Forestry and Grassland Administration and Ministry of Agriculture and Rural Affairs of P.R. China). Sampling was under the permission and guideline of Lanzhou University and Minqin Desert Botanical Garden.
Disclosure statement
The authors declare that they have no conflict interests.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MW927506. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA739272, SRS9240871, and SAMN19778433 respectively.
Additional information
Funding
References
- Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 1(5):2320–2325.
- Carreira VP, Padró J, Koch NM, Fontanarrosa P, Alonso I, Soto IM. 2014. Nutritional composition of Opuntia sulphurea G. Don Cladodes. Haseltonia. 19(19):38–45.
- Casas A, Barbera G. 2002. Mesoamerican domestication and diffusion. Cacti: biology and Uses. 143:62.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucl Acids Res. 45(4):e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Köhler M, Reginato M, Souza-Chies TT, Majure LC. 2020. Insights into chloroplast genome evolution across opuntioideae (Cactaceae) reveals robust yet sometimes conflicting phylogenetic topologies. Front Plant Sci. 11:729.
- Panico AM, Cardile V, Garufi F, Puglia C, Bonina F, Ronsisvalle S. 2007. Effect of hyaluronic acid and polysaccharides from Opuntia ficus indica (L.) cladodes on the metabolism of human chondrocyte cultures. J Ethnopharmacol. 111(2):315–321.
- Pimienta-Barrios E. 1994. Prickly pear (Opuntia spp.): a valuable fruit crop for the semi-arid lands of Mexico. J. Arid Environ. 28(1):1–11.
- Qu X, Moore RJ, Li D, Yi T. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.