Abstract
Campsosternus auratus (Drury, 1773) is a large, beautiful click beetle found in Southern China. The complete mitogenome of C. auratus was reported in this study. The mitogenome contains 15,943 base pairs. The composition of mitogenome is 39.6% for A, 9.9% for G, 32.8% for T, and 17.7% for C. A set of 37 genes, including 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and one control region, were annotated. The phylogenetic result supports the monophyly of Elateridae.
Click beetles (Elateridae), the ninth most diverse family of beetles, include more than 12,000 described species worldwide (Johnson Citation2002), of which most are pests that can damage crops and trees having the potential to adversely impact the production of agriculture and forestry. All Campsosternus species are large in size and exhibit a metallic luster on their body surface, which makes them appear similar to jewel beetles (Chrysochroa). There are about 17 species and subspecies of genus Campsosternus reported in China. C. auratus is a dominant species widespread in Southern China. It is a typical species of genus Campsosternus (Jiang Citation1991), usually can be found on firs (Cunninghamia lanceolata (Lamb.)), grassland, or under road lamp (Jiang Citation2002).
Three adults C. auratus specimens were collected in May 2021 from Guangzhou, China (N23°9′40″, E113°22′2″). The specimens were deposited into absolute ethanol in the Insect Collection at Qinghai Normal University, Xining, China (please contact Qingbai Hou, email: [email protected]) under the voucher number QNU2021C000063. Total genomic DNA was extracted from the thorax muscles of a single individual using the TIANGEN Genomic DNA Extraction Kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. Four pairs of overlapped PCR primers were designed to amplify the complete mitogenome of C. auratus. The KOD-Plus (http://www.bio-toyobo.cn/product_detail_13.html) was used for amplification following the procedures: the initial denaturation step was performed at 95 °C for 3 mins, followed by 36 cycles reaction of 15 s at 96 °C, annealing step at 50 °C for 30 s, elongation for 3 mins at 68 °C, and the final elongation step for 5 mins at 68 °C. Then, the PCR products of amplification were collected and mixed for Next Generation Sequencing. The mitogenome was sequenced by Illumina HiSeq-PE150 with 150 bps paired-end reads at Sangon Biotech Co., Ltd., Shanghai, China. The reads were assembled with SPAdes version 3.14.1 (Bankevich et al. Citation2012) and GetOrganelle (Jin et al. Citation2020). Then the assembled mitogenome was annotated by using the MITOs WebServer (http://mitos2.bioinf.uni-leipzig.de/index.py), and manual confirmation method with reference to other Elateridae species.
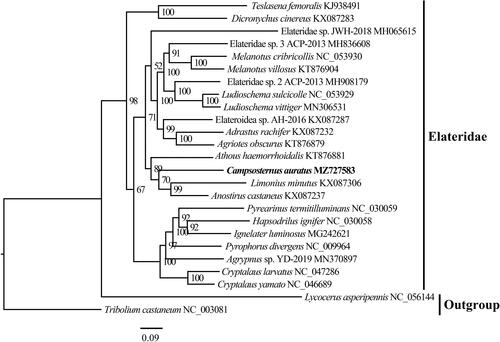
The complete mitogenome of C. auratus is a circular DNA molecule with a length of 15,943 base pairs, and contains 37 genes, including 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and one control region. Its sequence was deposited in GenBank under the accession number MZ727583. The phylogenetic relationship was reconstructed based on the 13 PCGs from 25 species using the IQ-tree on XSEDE with the maximum likelihood method (). The result highly supported the monophyly of Elateridae and showed that C. auratus was the sister group of Anostirus castaneus and Limonius minutus, which is consistent with the result based on 28S rDNA data (Reiko et al. Citation2007).
Figure 1. IQ-tree with the maximum likelihood method was constructed using 13 PCG sequence of C. auratus with 24 species of Coleoptera. The nodal numbers indicate the posterior possibility. Genbank accession numbers for the sequences are indicated next to the species names. The newly sequenced species are indicated in bold.
Ethical approval statement
This study was approved by the Administration Committee of Experimental Animals, Qinghai Province, China.
Authors contributions
Xiaoning Zhang was involved in the conception and design. Qingbai Hou was involved in the analysis and interpretation of the data. All authors contributed to the final manuscript and agree to the final approval of the version to be published. All authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ727583. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA754357, SRR15447068, and SAMN20769340 respectively.
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Jiang SH. 1991. Notes on the Chinese click beetles of the genus Campsosternus Latreille (coleoptera: elateridae), with discretions of two new species. Entomotaxonomia. 8 (5):275–279.
- Jiang SH. 2002. The dominant species in the Chinese elateridae fauna and the related questions. J Central South For Univ. 22 (4):55–60.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Johnson PJ. 2002. Elateridae. In: Arnett RH, Thomas MC, Skelley PE, Frank JH, editors. American beetles. Vol. 2. Boca Raton (FL): CRC Press; p. 160–173.
- Reiko SO, Yuichi O, Hitoo Ô. 2007. Phylogenetic relationships of click beetles (Coleoptera: Elateridae) inferred from 28S ribosomal DNA: insights into the evolution of bioluminescence in Elateridae. Mol Phylogenet Evol. 42(2):410–421.
