Abstract
Medicago scutellata (Linnaeus, 1753) is one of the essential leguminous forages distributed across tropical and subtropical regions of the world. In our study, we obtained the complete chloroplast genome of M. scutellata with a length of 124,082 bp. The total GC content of the entire chloroplast genome of M. scutellata was 33.9%. Among the 110 unique genes in the circular genome, 30 tRNA, 4 rRNA, and 76 protein-coding genes were successfully annotated. A phylogenetic tree constructed using common protein-coding genes revealed that M. scutellata is closely related to M. truncatula from the Fabaceae family.
Medicago scutellata (Linnaeus, 1753) is a major annual legume that is widely cultivated throughout the world, particularly in the Mediterranean region. It is a highly productive forage crop that is also stress tolerant, making it an excellent source and valuable forage crop for livestock fodder (Khaef and Sadeghi Citation2011). In addition, the seeds of M. scutellata also contain significantly higher trypsin inhibitor levels than other Medicago species, suggesting that M. scutellata could act as a potentiator of cisplatin-mediated trypsin inhibitor as a plant-derived anticancer agent for human breast and cervical cancers (Lanza et al. Citation2004). It also possesses extensive glandular stems and leaf hairs that serve as an anthelmintic mechanism and provide resistance against insects (Bauchan Citation1987).
Chloroplast is a vital organelle that has its own DNA genome and plays an important role in the fixation of carbon dioxide and the regulation of several metabolic pathways. The chloroplast genome of plants has been a focus of research in plant molecular evolutionary and phylogenetic studies (Clegg et al. Citation1994). In addition, the availability of the complete chloroplast genome sequences can facilitate the development of chloroplast transformation technology (Bock Citation2001). Therefore, the chloroplast genome needs to be investigated and analyzed in greater depth. However, so far, the chloroplast genome sequence of M. scutellata has not been reported. In the present study, the chloroplast genes of M. scutellata were sequenced and structurally characterized, providing a valuable resource for further studies on the genetic evolution of legumes, especially fodder crops. The chloroplast genome sequencing of M. scutellata will serve as a solid basis for molecular biology, genetic breeding of M. scutellate, and the development of new fodder crops (Tao et al. Citation2017).
The sample of M. scutellata was collected from the Bajia Botanical Garden (E116°29′, N40°03′) in Beijing, China. Seeds of M. scutellata were kept in the Forage Germplasm Bank of the School of Grassland Science, Beijing Forestry University (BFU) for conservation and available for use in research. And the voucher specimen were deposited in the Herbarium of Beijing Forestry University (http://cxy.bjfu.edu.cn/, Tiejun Zhang, [email protected]) under the voucher number MS535644. In the laboratory, leaves were collected from germinated plants, and genomic DNA was extracted using a DNA extraction kit from Shanghai Limin Industrial Co., Ltd (Shanghai, China). Sequencing was performed on the Illumina Novaseq PE150 platform (Illumina Inc, San Diego, USA) to generate 150 bp paired-end reads. The cleaned reads were assembled into complete chloroplast genomes using the software GetOrganelle v1.5 (Jin et al. Citation2018), using the previously described chloroplast genome of Medicago truncatula (GenBank accession number: NC_003119) as a reference. Annotation of the chloroplast genome was performed using the online programs CPGAVAS2 (Shi et al. Citation2019) and GeSeq (Tillich et al. Citation2017), followed by manual correction. The annotated chloroplast genome sequences were registered in GenBank under the accession number MZ895077. The study on M. scutellata, including the collection of plant material was carried out in accordance with guidelines provided by School of Grassland Science, Beijing Forestry University and Chinese or international regulations. Field studies has followed all the research protocols and complied with the legislation of Beijing.
In this study, the complete chloroplast genome of M. scutellata was determined to be 124,082 bp in length. The GC content of the entire chloroplast genome was 33.9%. A total of 110 genes were identified in the chloroplast genome, including 76 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Among them, 30 genes encoding amino acid transfer proteins, 15 genes encoding light-harvesting structural proteins of PSII, 11 genes encoding NADH dehydrogenase proteins, 11 genes encoding ribosomal proteins of the small subuni, were found in the chloroplast genome of M. scutellata.
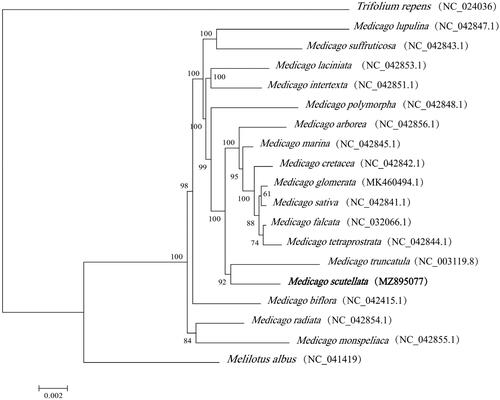
The chloroplast genomes of 17 species from the Fabaceae family and Melilotus albus and Trifolium repens as outgroup species were downloaded from the National Center for Biotechnology Information (NCBI) GenBank database to determine the phylogenetic relationships of M. scutellata. These sequences were aligned using MAFFT v7 (Katoh et al. Citation2019). In addition, a maximum likelihood (ML) tree based on the common protein-coding genes of 19 species was constructed using raxmlGUI1.5b (v8.2.10) (Silvestro and Michalak Citation2012). According to the results of the phylogenetic study, the entire tree was divided into three main clades. Two outgroup specie, Melilotus albus and Trifolium repens, formed two independent branches. Seventeen species from the Medicago group belonged to one clade. And M. truncatula was shown to be closely related to M. scutellata (). This study will provide valuable markers for species identification, phylogenetic relationships, and population genetics in the Fabaceae family, particularly for legume forage, as well as for other applications.
Figure 1. Phylogenetic tree reconstructed by the maximum likelihood (ML) method, based on common protein‐coding genes of 19 species of the Fabaceae family, with Melilotus albus and Trifolium repens, the sister group of Medicago within Fabaceae as outgroup. The numbers on the lines represent ML bootstrap values (>70%).
Author contributions
TZ was involved in the conception, design and the final approval of the version to be published. Both XH and YJ analyzed and interpreted the data, and XH drafted the paper. YS revised the paper critically for intellectual content, and was involved in the analysis of the data. All authors agree to be accountable for all aspects of the work.
Acknowledgements
We thank Yuehui Chao for his help in writing assistance.
Disclosure statement
The authors declare that they have no conflicts of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ895077. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA743404, SRR15390011, and SAMN19988356, respectively
Additional information
Funding
References
- Bauchan GR. 1987. Embryo culture of Medicago scutellata and M. sativa. Plant Cell Tiss Organ Cult. 10(1):21–29.
- Bock R. 2001. Transgenic plastids in basic research and plant biotechnology. J Mol Biol. 312(3):425–438.
- Clegg MT, Gaut BS, Learn GH, Morton BR. 1994. Rates and patterns of chloroplast DNA evolution. Proc Natl Acad Sci USA. 91(15):6795–6801.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 21:241.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Khaef N, Sadeghi H. 2011. Improving Germination rate of Medicago Scutellata and Medicago Rigidula as affected by seed-coat dormancy breaking techniques. Legume Res. 34(2):98–104.
- Lanza A, Tava A, Catalano M, Ragona L, Singuaroli I, Cuna FSRD, Cuna GRD. 2004. Effects of the Medicago scutellata trypsin inhibitor (MsTI) on cisplatin-induced cytotoxicity in human breast and cervical cancer cells. Anticancer Res. 24(1):227–233.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyser. Nucleic Acids Res. 47:65–73.
- Silvestro D, Michalak I. 2012. RaxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12(4):335–337.
- Tao X, Ma L, Zhang Z, Liu W, Liu Z. 2017. Characterization of the complete chloroplast genome of alfalfa (Medicago sativa) (Leguminosae). Gene Reports. 6:67–73.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes . Nucleic Acids Res. 45(W1):W6–W11.
