Abstract
The genus Dendronereis Peters, 1854 is characterized in the polychaete family Nereididae by its feather-shaped branchiae on the anterior segments. In this study, we present the first complete mitogenome of Dendronereis, represented by D. chipolini Hsueh, Citation2019, collected from Beibu Gulf, China. The nucleotide composition is biased toward A + T nucleotides, accounting 31.5% for A, 22.3% for C, 14.7% for G and 31.5% for T. The assembled mitogenome is 15,763 bp in length, with a typical set of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA), 2 ribosomal RNA (rRNA), and 1 non-coding control region. All genes are encoded on H-strand. The control region is 1260 bp in length and located between tRNA-Gly and tRNA-Met. Phylogenetic study showed that D. chipolini is arranged with high support into the clade of Namanereidinae. The complete mitogenome provides important molecular data for investigating the phylogeny and evolution of the nereid animals.
Polychaete are very common and frequently dominant group of marine organisms present from seashore to deep sea and recognized as a diverse, abundant and significant functional component of marine ecosystem (Misra Citation1999). Nereididae is one of the most diverse polychaete families with a wide distribution (Read and Fauchald Citation2021). Nereidids are utilized as the best-known polychaete bait for recreational fishers in Mediterranean Sea, and the Pacific coasts (especially east Asia) (Cole et al. Citation2018). Species classified to the genus Dendronereis are unique in the family Nereididae for their bipinnate branchiae, which are present on the anterior segments as a subdivision of the dorsal cirri. To date, five species are recognized within the genus Dendronereis (Hsueh Citation2019).
Samples of Dendronereis chipolini were collected from the estuary region of Maowei Sea, Beibu Gulf, China (21.9005°N, 108.6223°E) and then preserved in 100% ethanol. They were identified as Dendronereis chipolini Hsueh, Citation2019 based on a combination of characters such as lack of pharyngeal paragnaths, 7–8 pairs of branchiae starting from chaetiger 15, and homogomph spinigers with blunt-tipped short blades present in the neuropodia of chaetigers 3–12. Voucher specimens (voucher number BBGU-MP 010012, contact Dr. Wenquan Zhen, E-mail: [email protected].) were deposited in the herbarium of Beibu Gulf University. The insert size of the paired-end reads 350 bp long sequences were constructed by using a TruSeq DNA PCR-free library preparation kit. The Illumina Nova 6000 platform was used to sequence the mitochondrial genome, and 20,459,259 reads are assembled with NOVOPlasty 4.0 (Dierckxsens et al. Citation2017). Gene annotation was obtained using MITOS WebServer (Bernt et al. Citation2013) and tRNAscan-SE 2.0 (Lowe and Chan Citation2016; Chan and Lowe Citation2019).
The complete mitochondrial genome of D. chipolini is 15,763 bp in size (accession number: MW532084) and its overall base composition is 31.5% for A, 22.3% for C, 14.7% for G and 31.5% for T. The complete mitogenome of D. chipolini shows 75.96% and 73.33% identities to Platynereis dumerilii (AF178678) and Namalycastis abiuma (KU351089), respectively. The complete mitogenome of D. chipolini includes 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA), 2 ribosomal RNA (rRNA) and 1 non-coding control region. All PCGs, tRNA and rRNA genes were encoded on H-strand. The size of small ribosomal RNA (12S rRNA) and large ribosomal RNA (16S rRNA) genes is 826 bp and 1263 bp, respectively. The predicted D-loop region is 1260 bp in length and located between tRNA-Gly and tRNA-Met.
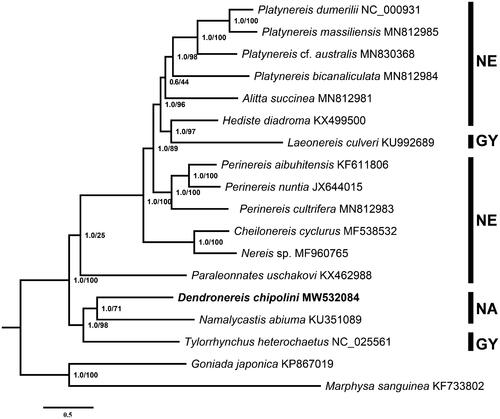
Phylogenetic trees of the family Nereididae were reconstructed based on the concatenated dataset of 13 mitochondrial PCGs of D. chipolini and 16 published polychaete species. Maximum likelihood phylogenies were inferred using IQ-TREE 1 (Nguyen et al. Citation2015). Bayesian Inference phylogenies were inferred using MrBayes 3.2.6 (Ronquist et al. Citation2012). Both the ML (Maximum Likelihood) and BI (Bayesian Inference) trees were consistent in topology, with high support values for most branches (). According to this analysis, the Nereididae is divided into two major clades. One of the clades consisted of species with pharyngeal paragnaths which are placed in the subfamily Nereidinae, and the species Laeonereis culveri which is classified in the subfamily Gymnonereidinae. The other one contains the species without paragnaths, which are classified in the subfamilies Namanereidinae and Gymnonereidinae. This analysis is basically consistent with previous research (Alves et al. Citation2020; Gomes-dos-Santos et al. Citation2021). The complete mitogenome of D. chipolini will provide valuable information to understand phylogenetic relationships among the marine polychaetes, especially the family Nereididae.
Figure 1. Bayesian inference (BI) tree of Nereididae reconstructed based on 13 mitochondrial PCGs. Numbers at the nodes represent BI posterior probability(left)/ML bootstrap scores (right). NE, Nereidinae; GY, Gymnonereidinae; NA, Namanereidinae. New sequence of the species D. chipolini is set in bold.
Disclosure statement
No potential conflict of interest was reported by the authors. The authors alone are responsible for the content and writing of this article.
This study did not need specific permissions or licenses for the studied animals belong to non-regulated invertebrates, which are collected from the mudflats.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession number MW532084. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA786757, SRR17153915, and SAMN23720049 respectively.
The research was conceived and designed by Dr Wenquan Zhen, Pro Erwei Hao, Pro Jiagang Deng, Pro Junhua Zhu and Pro Youhou Xu. Sampling and mitochondrial genome data analysis were conducted by Dr Wenquan Zhen and Miss Wanru Xu. Species identification and molecular phylogenetic analysis were performed by Dr. Xuwen Wu. The drafting and revising of the paper were conducted by Dr Wenquan Zhen, Xuwen Wu and Pro Youhou Xu. All authors agree to be accountable for all aspects of the work.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Alves PR, Halanych KM, Santos CSG. 2020. The phylogeny of Nereididae (Annelida) based on mitochondrial genomes. Zool Scr. 49(3):366–378.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14.
- Cole VJ, Chick RC, Hutchings PA. 2018. A review of global fisheries for polychaete worms as a resource for recreational fishers: diversity, sustainability and research needs. Rev Fish Biol Fisheries. 28(3):543–565.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Gomes-dos-Santos A, Hagemann A, Valente L, Malzahn AM, Monroig Ó, Froufe E, Castro LFC. 2021. Complete mitochondrial genome of the ragworm annelid Hediste diversicolor (of Müller, 1776) (Annelida: Nereididae). Mitochondrial DNA Part B. 6(10):2849–2851.
- Hsueh P-W. 2019. Two new species of nereidids (Annelida, Polychaeta) from Taiwan. Zootaxa. 4652(3):zootaxa.4652.3.10.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–57.
- Misra A. 1999. Polychaeta. In: Goshe AK, editor. Fauna of West Bengal – part 10. Calcutta: Zoological Survey of Indica; p. 125–225.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Read G, Fauchald K. 2021. World Polychaeta database. Nereididae Blainville, 1818. World Register of Marine Species; [accessed 2021 Jan 20]. http://www.marinespecies.org/aphia.php?p=taxdetails&id=22496
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
