Abstract
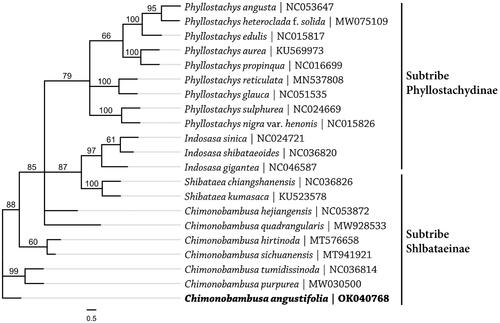
Chimonobambusa angustifolia is a famous ornamental and edible bamboo species. Here, the complete chloroplast (cp) genome of C. angustifolia was assembled and annotated. The cp genome is 139,611 bp in size, consisting of two copies of inverted repeat (IR) regions of 21,799 bp, one large single-copy (LSC) region of 83,202 bp, and one small single-copy (SSC) region of 12,811 bp. It encodes 133 genes (110 unique), including 86 protein-coding genes (77 unique), 39 tRNA genes (29 unique), and 8 rRNA genes (4 unique). Phylogenetic analysis based on 21 cp genome sequences within four genera of family Poaceae indicated that genus Chimonobambusa were closely related to genus Shibataea, both belong to subtribe Shlbataeinae.
Chimonobambusa angustifolia C. D. Chu et C. S. Chao 1981 is one kind of bamboo. Its culm is 2–5 m tall, with diameter 1–2.5 cm. It is distributed in Guangxi, Guizhou, Hubei and Shanxi provinces in China, with an altitude range of 700–1400 m (Li and Chris, Citation2006). Its new shoot grows from August to September and can be used as food. Due to the presence of straight and hard culm, it is widely used for papermaking and bamboo weaving. The nutritional components of bamboo shoots among C. angustifolia, C. hejiangensis and C. utilis are very close (Guo et al. Citation2010), which all are the staple food bamboos for giant panda and red panda (Nijboer and Dierenfeld, Citation2011; Shi et al. Citation2020). C. angustifolia has good economic value, and its planting is considered to be one of the main forestry industries for poverty alleviation (Zhang et al. Citation2017). In this study, we report and characterize the chloroplast (cp) genome of C. angustifolia.
Fresh and healthy leaves of a single plant of C. angustifolia were collected from Jiuba town, Tongzi County, Zunyi City, Guizhou Province (N 28°13′30″, E 106°42′38″, altitude: 1460 m). The voucher specimen (Wu20210053) was deposited in the Herbarium of Yunnan Academy of Forestry and Grassland, Kunming, China (http://www.ynlky.org.cn/, Wu Tao, [email protected]). The total genomic DNA was isolated using DNAsecure Plant Kit (Beijing Tiangen Biotech Company). The concentration and integrity of the DNA were analyzed by Invitrogen Qubit Fluorometers (Thermo Fisher Scientific) and agarose gel electrophoresis. DNA library were sequenced on the Hiseq 2500 (Illumina, CA, USA) with 150 bp paired ends, 2.3 Gb reads were obtained and then were filtered using NGS QC Toolkit_v2.3.3 (Patel and Jain Citation2012). The cp genome assembly was performed using GetOrganelle (Jin et al. Citation2020) and was annotated using Geneious Prime 2020.0.5 (Kearse et al. Citation2012) with C. hejiangensis chloroplast genomes (NC053872) as references. The assembled and annotated cp genome of C. angustifolia was deposited at GenBank under the accession number OK040768.
The complete cp genome of C. angustifolia is 139,611 bp in size, consisting of one large single-copy (LSC) region (83,202 bp), one small single-copy (SSC) region (12,811 bp), and two copies of inverted repeat (IR) regions (21,799 bp). The entire GC content of C. angustifolia is 38.90%, and the ones of the LSC, SSC, and IR regions are 36.98%, 33.23%, and 44.22%, respectively. The genome contained 133 genes (110 unique), including 86 protein-coding genes (77 unique), 39 tRNA genes (29 unique), and 8 rRNA genes (4 unique). The complete cp genome sequences of 21 species from four genera, Indosasa, Phyllostachys, Chimonobambusa, Shibataea, in the tribe Shlbataeeae of family Poaceae (Gramineae) were aligned using MAFFT (Katoh and Standley Citation2013) in order to ascertain the phylogenetic status of C. angustifolia. A maximum likelihood tree was reconstructed with RAxML8 (Stamatakis Citation2014), which revealed that genus Chimonobambusa were closely related to genus Shibataea, both belong to subtribe Shlbataeinae (). The newly characterized complete chloroplast genome of C. angustifolia will provide essential resources for further study on the evolution and genetic diversity.
Author contributions
T.X. and T.W. conceived and designed the research; T.X. and Y.L. collected the samples, performed the experiments and analyzed the data; T.W. and T.X. wrote the drafting of the paper and revised the manuscript. All authors approved the final manuscript and agreed to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
– The complete chloroplast genome generated for this study has been deposited in GenBank with accession number OK040768, which is openly available in GenBank of NCBI at website (https://www.ncbi.nlm.nih.gov/). All high-throughput sequencing data files are available from the GenBank Sequence Read Archive (SRA) accession number: SRR15725929. The associated BioProject and Bio-Sample numbers are PRJNA760964 and SAMN21240066 respectively.
Additional information
Funding
References
- Guo GQ, Ding YL, Yang L, et al. 2010. Nutrient analysis on bamboo shoots of three species in Chimonobambusa. China Vegetables. 16:79–81.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li DZ, Chris S, et al. 2006. Chimonobambusa. In: Shouliang C, Dezhu L, Guanghua Z, editors. Flora of China, Vol. 22: Poaceae. Beijing: Science Press; p. 152–161.
- Nijboer J, Dierenfeld ES. 2011. Red panda nutrition: how to feed a vegetarian carnivore. In Glatston AR, editors. Red panda—biology and conservation of the first panda. London: Academic Press; p. 257–270.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLOS One. 7(2):e30619.
- Shi J, Chen Q, Huang J, et al. 2020. Biodiversity of the staple food bamboos of giant panda and its important value. World Bamboo Rattan. 18(5):10–19.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Zhang D, Xu J, He T, et al. 2017. Type division and application evaluation of Chimonobambusa angustifolia forest community in Meitan County. Guizhou Province. World Bamboo and Rattan. 15(6):6–12.