Abstract
Pleione hookeriana (Lindl.) B. S. Williams is a species of Orchidaceae with high ornamental value. It is a protected plant in China. To document the genetic history of this rare species, the chloroplast genome sequence of P. hookeriana from the Yunnan Province, China, were analyzed. The complete chloroplast genome is 158,930 bp in length and contains a large single-copy (LSC) region of 86,880 bp, a small single-copy region of 18,664 bp (SSC), and a pair of inverted repeats (IR) of 26,693bp. There are 137 genes, including 89 protein-coding, 40 transfer RNA (tRNA) and 8 ribosomal RNA (rRNA) genes. The total GC content of the chloroplast genome sequence is 37.2%. The maximum-likelihood phylogenetic analysis indicated that P. hookeriana was sister to P. chunii (MK792342.1). The result may be because the species are roughly the same geographical location and advanced and developed from the same ancestor. This study provides important information for the identification and conservation of species, and genetic engineering of P. hookeriana.
Pleione hookeriana (Lindl.) B. S. Williams (Pleione hookeriana (Lindl.), Rollisson, William 1875) is an epiphytic herb that grows on tree trunks and moss-covered rocks or rock walls at the edge of shrubs. Distributed in the north of Guangdong, the west to the north of Guangxi, the southeast of Guizhou, the southeast of Yunnan and the south of Tibet between 1600 and 3100 meters above sea level. Due to the lack of extensive sampling and sufficient molecular evidence, the classification of P.hookeriana and P. chunii is still in a fuzzy state (Zhang et al. Citation2018). In recent years, the comparative analysis of the complete chloroplast genomes of different related species has provided promising methods for the study of phylogeny, population dynamics, and species evolution (Shaw et al. Citation2014; Jiang et al. Citation2019).
The research content and data reported in this paper are all conducted ethically and comply with relevant experimental and legislative guidelines. The collection of plant materials involved and the acquisition of data have been ethically approved by local agencies. Fresh leaves tissue of P. hookeriana were collected from Heqing County, Dali City, Yunnan Province (26°28'35″N, 100°25'32"E). The voucher specimen was preserved at HAAS (Hangzhou Academy of Agricultural Sciences, http://www.hznky.com) in charge of Weiguo Chai (Voucher number: Weiguo Chai et al 2103005 and [email protected]).
Total genomic DNA was extracted using a modified CTAB method. The DNA was deposited at the Key Laboratory of Plant Secondary Metabolism and Regulation in Zhejiang Province, Zhejiang Sci-Tech University (http://sky.zstu.edu.cn) under the voucher number ZSTUX0201 (collected by Pengguo Xia and [email protected]). The sequencing library was constructed by the Illumina HiseqTM platform with an insert size of about 400 bp. In total, Raw PE reads were generated and approximately 2.508 Gb of clean PE reads were obtained after removing adapter and low-quality bases. NOVOPlasty v2.7.2 (Dierckxsens et al. Citation2017) was used to assemble the complete chloroplast genome of P. hookeriana with default settings. The sequence was annotated using geneious Prime. The complete chloroplast genome of P. hookeriana was obtained and submitted to GenBank (Accession number MZ958823). The complete chloroplast genome sequence of P. hookeriana is 158,930 bp in length and contains a LSC of 86,880 bp, a SSC of 18,664 bp and a IRs of 26,693 bp. There are 137 genes, including 89 protein-coding, 40 transfer RNA (tRNA) and 8 ribosomal RNA (rRNA) genes. The total GC content of the chloroplast genome sequence is 37.2%. Compared with the structures of P. pleionoides, P. forrestii, P. chunii, P. maculata and P. formosana, P. hookeriana has two more tRNAs. The GC content of genus Pleione are 37.3% except P. maculata, which is 37.2% (Jiang et al Citation2019; Chen et al Citation2019; Wu, Shen, et al Citation2019; Wu, Wu, et al Citation2019; He et al Citation2021).
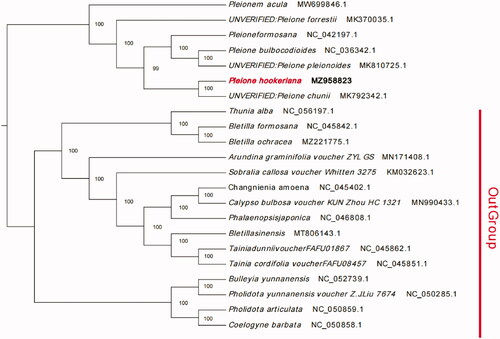
The phylogenetic analysis of the P. hookeriana complete chloroplast genome and 21 related species was constructed using the best-selected TVM + F+R4 model and 1000 bootstrap replicates with IQTREE V1.6.7 (Nguyen et al.Citation2015). The maximum-likelihood phylogenetic analysis indicated that P. hookeriana was sister to P. chunii (MK792342.1) (). P. chunii is regarded by some authorities as a variant of P. hookeriana. In fact, they are all epiphytic plants with very similar lip shapes and flower colors. However, P. chunii has obvious pseudobulbs, larger flowers, and callus on the lips (). The maximum-likelihood phylogenetic analysis indicated that P. chunii was sister to P. bulbocodioides and P. formosana (Wu SS et al. Citation2019). A similar situation appeared in our evolutionary analysis, from which we can conclude that P. hookeriana was sister to P. chunii (MK792342.1).
Author contributions
W.C. and P.X. conceived and designed this study. X. C. and H. L. conducted analysis. P.X. and X.C. contributed the analytical methods. W.C. wrote the manuscript. P.X. edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Ethical approval
Research and collection of plant material was conducted according to the guidelines provided by HAAS. Permission was granted by Hangzhou Academy of Agricultural Sciences to carry out research on the species.
Disclosure statement
The authors report no conflicts of interests.
Data availability statement
The data that support the findings of this study are openly available in NCBI (https://www.ncbi.nlm.nih.gov) GenBank with the accession number (MZ958823). The associated BioProject, SRA and BioSample numbers are PRJNA755890, SRR15524497, and SAMN20856062 respectively.
Additional information
Funding
References
- Chen JL, Dai ZW, Dai XY, Lan SR, Wu SS. 2019. The complete chloroplast genome of Pleione pleionoides (Orchidaceae). Mitochondrial DNA Part B. 4(2):2167–2168.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- He LF, Qiang SJ, Zhang YH. 2021. The complete chloroplast genome of Pleione maculata, an orchid with important ornamental value and medicinal value. Mitochondrial DNA Part B. 6(8):2263–2265.
- Jiang MT, Chao WC, Huang CL, Lan SR, Liu ZJ, Wu SS. 2019. The complete chloroplast genome of Pleione formosana (Orchidaceae). Mitochondrial DNA Part B. 4:1–3.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Shaw J, Shafer HL, Leonard OR, Kovach MJ, Schorr M, Morris AB. 2014. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: the tortoise and the hare IV. Am J Bot. 101(11):1987–2004.
- Wu SS, Shen LM, Ling R, Dai ZW, Liu ZJ, Lan SR. 2019. Next-generation sequencing yields the complete chloroplast genome of Pleione chunii, a vulnerable orchid in China. Mitochondrial DNA Part B. 4(2):2576–2578.
- Wu SS, Wu XQ, Dong N, Ling R, Jiang H. 2019. Next-generation sequencing yields the complete chloroplast genome of Pleione forrestii (Orchidaceae). Mitochondrial DNA Part B. 4(2):2777–2778.
- Zhang WEI, Qin JIAO, Yang RUI, Yang YI, Zhang S-B. 2018. Two new natural hybrids in the genus Pleione (Orchidaceae) from China. Phytotaxa. 350(3):247–258.