Abstract
Rubus ellipticus Sm. var. obcordatus Focke is an important species in the phylogeny and evolution of genus Rubus L. in the family Rosaceae. Its chloroplast genome, as reported in this study, is 155,656 bp in size, and it has an average GC content of 37.14%. The chloroplast genome showed a typical quadripartite structure comprising a large single copy (LSC) region (85,388 bp) and a small single copy (SSC) region (18,730 bp), which were separated by a pair of inverted repeats (IRs, 25,769 bp). In total, this plastome was found to contain 129 different genes, including 85 protein-coding genes, 36 tRNA genes, and eight rRNA genes. The completed chloroplast genome of R. ellipticus var. obcordatus will set a new insight into clarifying the phylogeny and genomic studies in genus Rubus of the family Rosaceae.
Rubus ellipticus Sm. var. obcordatus Focke belongs to genus Rubus L. (Rosaceae) section Idaeobatus Focke, subsection Stimulantes Yu et Lu. Its fresh golden fruits are seasonally used for food, and its twigs and leaves are frequently used for medicine by indigenous people in southwestern of China. It is extremely difficult to distinguish the species in genus Rubus from each other. This is because of their complex morphological variations (Alice and Campbell Citation1999) due to their inter- and intraspecific hybridization, polyploidization, and apomixis, especially between R. ellipticus Sm. var. ellipticus (its original variant) and R. pinfaensis H. Lév. & Vaniot. To establish the relationships between different species clearly, it is necessary to rebuild our understanding of these plants’ phylogeny based on different molecular markers.
In plants, the chloroplast genome offers several advantages over the nuclear and mitochondrial genomes (Wolfe et al. Citation1987; Shendure and Ji Citation2008). It can provide plentiful polymorphisms to resolve taxonomic and phylogenetic discrepancies (Clegg and Zurawski Citation1992). The chloroplast genome has already been used in many plant phylogenies at different taxonomic levels and phylogenetic key nodes (Moore et al. Citation2010; Huang et al. Citation2017; Shen et al. Citation2017; Mehmood, Abdullah Shahzadi, Ahmed, et al. 2020; Mehmood, Abdullah Ubaid, Shahzadi, Ahmed, et al. 2020; Mehmood, Abdullah Ubaid, Shahzadi, Bao, et al. 2020), and has successfully resolved many problems in taxonomic and phylogenetic relationships in difficult taxa.
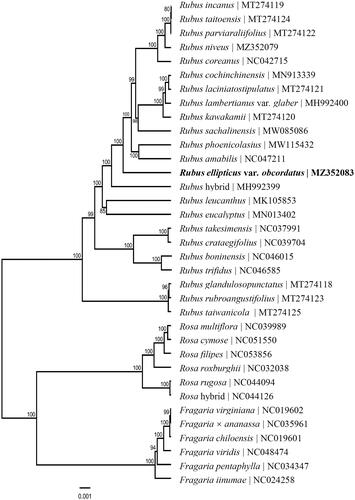
In this study, the complete chloroplast genome of R. ellipticus var. obcordatus is reported. This is a wild species widespread in tropical, subtropical, and temperate zones in Guangxi, Sichuan, Yunnan, and Guizhou Provinces of China (Robertson Citation1974; Yu and Lu Citation1985; Thompson Citation1995; Lu and Boufford Citation2003). It lives at elevations between 300 and 2000 m, as a molecular resource for future use on the taxonomy of Rubus. The fresh leaves of R. ellipticus var. obcordatus were collected from Cuihua Town, Daguan County, Zhaotong City, Yunnan Province (27°45′14′′ N, 103°53′51′′ E). The specimens were stored at the Herbarium of Horticultural Plants, Yunnan Agricultural University (https://www.ynau.edu.cn/, Shiyu Wang, [email protected]) under the voucher number Zhu-20201014R02. In order to construct chloroplast DNA libraries, total genomic DNA was extracted from fresh leaves using DNA Plantzol Reagent (Invitrogen, Carlsbad, CA). The extracted DNA was sequenced by Illumina HiSeq Sequencing System (Illumina, San Diego, CA) and a shotgun library was constructed. About 2.55 Gb pair-end (150 bp) raw reads were obtained, and the low-quality sequences were filtered using CLC Genomics Workbench version 8.0 (CLC Bio, Aarhus, Denmark) to produce high-quality clean reads. NOVOPlasty software (Dierckxsens et al. Citation2017) was used to align and assemble the complete chloroplast genome with the reference genome of R. niveus Thunb (MT576936). The complete chloroplast genome of R. ellipticus var. obcordatus was automatically annotated using CPGAVAS2 (Shi et al. Citation2019), and then adjusted and confirmed with Geneious version 9.1 (Kearse et al. Citation2012). The complete chloroplast genome was submitted to the GenBank database under the accession number of MZ352083. To ascertain the phylogenetic status of R. ellipticus var. obcordatus, the complete chloroplast genome sequences of 35 species from three genera (6 species from Fragaria L., 6 species from Rosa L., and 23 species from Rubus L.) in the family Rosaceae were aligned using MAFFT (Katoh and Standley Citation2013). With these sequence alignments, a maximum likelihood (ML) phylogenetic tree was reconstructed by RAxML8 (Stamatakis Citation2014) with GTR GAMMA I nucleotide model and 1000 bootstrap replicates.
The length of the chloroplast genome of R. ellipticus var. obcordatus is 155,656 bp, with an average GC content of 37.14%, which exhibited a typical quadripartite structure comprising a large single copy (LSC) region of 85,388 bp and a small single copy (SSC) region of 18,730 bp separated by a pair of identical inverted repeats (IRs) of 25,769 bp each. The chloroplast genome contains 129 genes (112 unique), including 85 protein-coding genes (79 unique), 36 tRNA genes (29 unique), and eight rRNA genes (4 unique).
The ML phylogenetic tree shows that 23 species of the genus Rubus formed an independent monophyly, with bootstrap support values of nearly 100% (), and 12 species in genera Fragaria and Rosa formed a monophyly with two clades. The monophyly of three genera of the family Rosaceae was found to be well-supported by the complete chloroplast genome sequence. This study lays the foundation for further understanding of the chloroplast genome information of Rubus plants, and provides a new insight into elucidating the phylogeny and genomics of the family Rosaceae.
Author contributions
Ying-an Zhu, Zheng-an Yang, and Jie Zhang were involved in the conception and design, Junjun Xie, and Guansong Yang analyzed and interpreted the data; Ying-an Zhu drafted the article, Shiyu Wang, Zheng-an Yang, and Jie Zhang revised it critically for intellectual content; all authors approved the final version to be published; and agreed to be accountable for all aspects of the work.
Disclosure statement
The authors have no potential or actual conflicts of interest to report. All authors promise that we have no any unethical behaviors for the study, the collections of plant materials were carried out in accordance with guidelines provided by the authors’ institution(s) and national or international regulations, and the field studies were complied with local legislation. If we have any unethical and illegal behaviors, we will take all responsibilities.
Data availability statement
The complete chloroplast genome generated for this study has been deposited in GenBank under accession number MZ352083, which is openly available in GenBank through the NCBI at website (https://www.ncbi.nlm.nih.gov/). All high-throughput sequencing data files are available from the GenBank Sequence Read Archive (SRA) under accession number: SRR14757446. The associated BioProject and Bio-Sample numbers are PRJNA735803 and SAMN19602746, respectively.
Additional information
Funding
References
- Alice L, Campbell C. 1999. Phylogeny of Rubus (Rosaceae) based on nuclear ribosomal DNA internal transcribed spacer region sequences. Am J Bot. 86(1):81–97.
- Clegg MT, Zurawski G. 1992. Chloroplast DNA and the study of plant phylogeny: present status and future prospects. In: Soltis PS, Soltis DE, Doyle JJ, editor. Molecular systematics of plants. Boston (MA): Springer; p. 1–13.
- Dierckxsens N, Mardulyn P, Smits G. 2017. Novoplasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Huang YY, Cho ST, Haryono M, Kuo CH. 2017. Complete chloroplast genome sequence of common bermudagrass (Cynodon dactylon (L.) Pers.) and comparative analysis within the family Poaceae. PLOS One. 12(6):e0179055.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lu LD, Boufford DE. 2003. Rubus linnaeus. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 9. Beijing, China: Science Press; St. Louis (MO): Missouri Botanical Garden Press; p. 195–285.
- Mehmood F, Abdullah Shahzadi I, Ahmed I, Waheed MT, Mirza B. 2020a. Characterization of Withania somnifera chloroplast genome and its comparison with other selected species of Solanaceae. Genomics. 112:1522–1530.
- Mehmood F, Abdullah Ubaid Z, Shahzadi I, Ahmed I, Waheed MT, Poczai P, Mirza B. 2020b. Plastid genomics of Nicotiana (Solanaceae): insights into molecular evolution, positive selection and the origin of the maternal genome of Aztec tobacco (Nicotiana rustica). PeerJ. 10:7717.
- Mehmood F, Abdullah Ubaid Z, Shahzadi I, Bao YM, Poczai P, Mirza B. 2020c. Comparative Plastomics of Ashwagandha (Withania, Solanaceae) and Identification of Mutational Hotspots for Barcoding Medicinal Plants. Plants. 9:752.
- Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of educations. Proc Natl Acad Sci USA. 107(10):4623–4628.
- Robertson K. 1974. The genera of Rosaceae in the southeastern United States. J Arnold Arboretum. 55:352–360.
- Shen XF, Wu ML, Liao BS, Liu ZX, Bai R, Xiao SM, Li XW, Zhang BL, Xu J, Chen SL. 2017. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules. 22(8):1330.
- Shendure J, Ji H. 2008. Next-generation DNA sequencing . Nat Biotechnol. 26(10):1135–1145.
- Shi LC, Chen HM, Jiang M, Wang LQ, Wu X, Huang LF, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–73.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Thompson M. 1995. Chromosome numbers of Rubus species at the National Clonal Germplasm Repository. HortSci. 30(7):1447–1452.
- Wolfe KH, Li WH, Sharp PM. 1987. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA. 84(24):9054–9058.
- Yu TT, Lu LT. 1985. Rubus Linnaeus. In: Yu TT, editor. Flora reipublicae popularis sinicae. Vol. 37. Beijing, China: Science Press; p. 10–218.