Abstract
Corylopsis sinensis Hemsl. is a deciduous shrub endemic to China, which is a valuable medicinal and ornamental species. In this study, we report the complete chloroplast genome sequence of C. sinensis, providing a genomic basis for future research. The chloroplast genome is 159,419 base pairs (bp) in length, with a large single-copy (LSC) region of 88,152 bp, a small single-copy (SSC) region of of 18,701 bp, and two inverted repeat (IR) regions of 26,283 bp. The overall GC content is 38.0% and the chloroplast genome encodes 113 unique genes including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The phylogenetic results show that C. sinensis is sister to C. spicata. These results of C. sinensis will improve our understanding of the evolution of Corylopsis and Hamamelidaceae.
Corylopsis sinensis Hemsl. is a deciduous shrub of the family Hamamelidaceae (Li et al. Citation1999). This species is native to China, and mainly distributed in forests or mountains of southern China (Zhang et al. Citation2003). C. sinensis is cultivated as an ornamental in gardens and parks due to its beautiful yellow flowers in early spring (Iwashina et al. Citation2009; Kim et al. Citation2016;). In addition, the whole plant of C. sinensis contains bergenin and it is a traditional Chinese medicine for the treatment of chronic bronchitis (Li et al. Citation2011). Therefore, C. sinensis has high ornamental and medicinal value. Furthermore, determining the systematic position of C. sinensis will provide valuable information for the evolution of Corylopsis and Hamamelidaceae. As a semi-autonomous organelle, chloroplast play significant role in photosynthesis and physiology of plant (Chang et al. Citation2021). However, the chloroplast genome of C. sinensis remains unpublished. Here, we assembled and characterized the complete chloroplast genome of C. sinensis to provide valuable genomic resources for further studies.
Fresh leaves of C. sinensis were collected from Chengdu Botanical Garden (104°7′42.54″E, 30°45′54.07″N), Sichuan province, China, and the specimen was deposited in Herbarium of Sichuan University (SZ) (http://mnh.scu.edu.cn/, Hong Chang, [email protected]) under the voucher number SZ02049686. The total genomic DNA was extracted using a modified CTAB method (Doyle and Doyle Citation1987) from about 0.2 g of leaves. The library with insert size of 300 bp fragments was prepared and sequenced by Illumina platform. The raw reads were first filtered to remove paired-end reads if either of the reads contained (i) adapter sequences, (ii) more than 10% of N bases, and (iii) more than 50% of bases with a Phred quality score less than five. The filtered reads were then assembled using NOVOPlasty version 2.7.2 (Dierckxsens et al. Citation2017). The assembled chloroplast genome was annotated using Plann version 1.1 (Huang and Cronk Citation2015). The positions of exons and introns of annotated genes were manually inspected and adjusted by Geneious version 11.0.3 (Kearse et al. Citation2012) and Sequin version 15.50. (http://www.ncbi.nlm.nih.gov/Sequin/). The annotated chloroplast genome was submitted to GenBank (accession number: MZ590567). To investigate the phylogenetic location of C. sinensis, the complete chloroplast genome sequences of C. sinensis and ten reported Hamamelidaceae species and two outgroups (Daphniphyllum pentandrum Hayata and Cercidiphyllum japonicum Siebold & Zucc.) were aligned using MAFFT version 7 software (Katoh and Standley Citation2013). IQ-tree software (Nguyen et al. Citation2015) was used to construct maximum likelihood (ML) tree under standard mode with 1000 bootstrap (Anisimova et al. Citation2011). The nucleotide substitution model was TVM + F+R2 according to the results of ModleFinder (Kalyaanamoorthy et al. Citation2017). The phylogenetic tree was visualized using online software ITOL V4 (https://itol.embl.de/; Letunic and Bork, Citation2019).
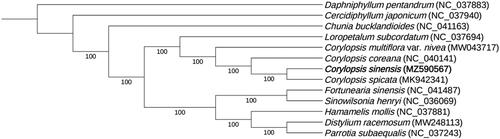
The complete chloroplast genome of C. sinensis is 159,419 base pairs (bp) in length and the GC content is 38.0%. It contains a large single-copy (LSC) region of 88,152 bp, a small single-copy (SSC) region of 18,701 bp, and two inverted repeat sequence (IR) regions of 26,283 bp. It encodes a total of 113 unique genes, including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The phylogenetic tree indicates that C. sinensis is sister to C. spicata, and the clade position of genus Corylopsis is the sister of genus Loropetalum with both a 100% bootstrap support (). The complete chloroplast genome of C. sinensis and the phylogenetic relationships will enhance our understanding of the evolution of Corylopsis and Hamamelidaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov) under the accession no. MZ590567. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA766356, SUB10430244, and SAMN21844338 respectively.
Additional information
Funding
References
- Anisimova M, Gil M, Dufayard J-F, Dessimoz C, Gascuel O. 2011. Survey of Branch Support Methods Demonstrates Accuracy, Power, and Robustness of Fast Likelihood-based Approximation Schemes. Syst Biol. 60(5):685–699.
- Chang H, Zhang L, Xie H, Liu J, Xi Z, Xu X. 2021. The conservation of chloroplast structure and improved resolution of infrafamilial relationships of Crassulaceae. Front Plant Sci. 12:631884.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A Rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15.
- Huang DI, Cronk QC. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47(W1):W256-W259.
- Iwashina T, Takemura T, Mishio T. 2009. Chalcone glycoside in the flowers of six Corylopsis as yellow pigment. J Japan Soc Hort Sci. 78(4):485–490.
- Kalyaanamoorthy S, Minh BQ, Wong T. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 1(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kim JH, Lee AK, Suh JK. 2016. Effect of warm and cold stratification, and ethanol treatment on germination of Corylopsis seeds. Hortic. Sci. 43(2):84–91.
- Li JH, Bogle AL, Klein AS. 1999. Phylogenetic relationship of Hamamelidaceae inferred from sequence of internal transcribed spacers (ITS) of nuclear ribosomal DNA. Am J Bot. 86(7):1027–1037.
- Li CS, Chen XZ, Fang DM, Li GY. 2011. A new bergenin derivative from Corylopsis willmottiae. Chem Nat Compd. 47(2):194–196.
- Nguyen LT, Schmidt H. v, Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Zhang ZY, Zhang HD, Endress P. 2003. HAMAMELIDACEAE. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 9. St. Louis: Science Press/Beijing: Missouri Botanical Garden Press. p. 18–42.