Abstract
Epimedium jinchengshanense Y. J. Zhang & J. Q. Li 2014 is an important ornamental and medicinal herb, but of unclear taxonomy. In this study, the complete chloroplast genome of E. jinchengshanense was sequenced. The genome was 157,169 bp in length, with a large single-copy region of 88, 520 bp, a small single-copy region of 17,075 bp and 2 inverted repeat regions of 25, 787 bp. The genome consisted of 113 unique genes, including 79 protein-coding genes, 30 tRNA genes and 4 rRNA genes. The GC contents were 38.78%. Phylogenetic analysis showed a close relationship between E. jinchengshanense and E. ilicifolium, which was explained by the morphological similarity of flowers and leaves of the two species.
1. Background
Epimedium L., the largest genus of Berberidaceae, is an herbaceous perennial with great economic value (Stearn Citation2002; Ying et al. Citation2011; Xu et al. Citation2019). Among the 68 published Epimedium species, 58 (85.3%) are distributed in China, and 57 are endemic to China (Xu et al. Citation2020a). The taxonomy and phylogenetic relationship within Epimedium remained controversial due to the large morphological variations of the genus (Liu et al. Citation2017; Xu et al. Citation2019, Citation2020a,Citationb; Guo et al. Citation2022). Although the taxonomy system described by Stearn (Citation2002) was globally accepted, 13 new species have been published in the last two decades (Xu et al. Citation2020a). Among them, Epimedium jinchengshanense Y. J. Zhang & J. Q. Li 2014, named after its type locality (Jinchengshan, Sichuan, China), was firstly described by Zhang et al. (Citation2014) due to its unique floral characteristics. The plant had been previously treated as E. wushanense. It was narrowly distributed in montane forests and thickets in northeastern Sichuan, at elevations of 600–1500 m (Zhang et al. Citation2014). Despite its narrow distribution, E. jinchengshanense has important medicinal and ornamental values because of its abundant medicinal components and large and showy flowers (Zhang et al. Citation2014; Jiang Citation2020). In this study, the complete chloroplast (cp) genome of E. jinchengshanense was sequenced and analyzed, which will facilitate the taxonomic research, resource protection and utilization of the valuable germplasm.
2. Methods
The sample of E. jinchengshanense was collected from the type locality, Jinchengshan, Yilong County, Sichuan Province, China (latitude 31.2100, longitude 109.9006). A specimen was deposited at the Herbaria of Jiangxi University of Chinese Medicine (JXCM; https://www.jxutcm.edu.cn/, Yanqin Xu, [email protected]) under the voucher number S. X. Liu & L. J. Liu 2016027. DNA was isolated from 0.3 g silica-dried leaf tissue using the CTAB protocol (Doyle and Doyle Citation1987). DNA libraries were prepared with an insert size of 350 bp using NEBNext Ultra DNA Library Prep Kit. Paired-end sequencing (150 bp reads) was performed on an Illumina NovaSeq 6000 platform (San Diego, CA) at the Novogene Co. Ltd. (Beijing, China). Finally, 5.83 G raw data was obtained with the number of raw reads 19,429,756. The cp genome was assembled using GetOrganelle v1.7.1 with default parameters (Jin et al. Citation2020), and the degree of coverage was 99.75%. The genes were annotated using GeSeq (Tillich et al., Citation2017) with E. ilicifolium (NC_044897) as a reference. The software Geneious primer (www.geneious.com) was used for further manual annotation with E. ilicifolium as a reference. The cp genome sequence of E. jinchengshanense was deposited in the GenBank database (MZ603799).
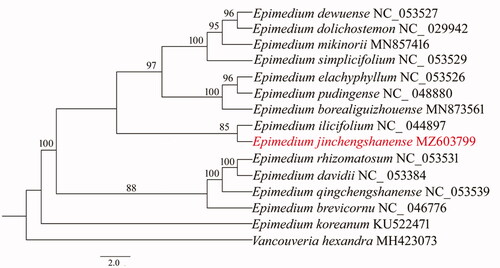
The phylogenetic relationship within Epimedium was reconstructed based on cp genomes of E. jinchengshanense and additional 14 Epimedium species previously reported, with Vancouveria hexandra (MH423073) as an out-group. Using PhyML v.3.0 (Stéphane et al. Citation2010), the maximum-likelihood (ML) tree was built with the best model of TVM + G and 1000 bootstraps.
3. Results
The complete chloroplast genome of E. jinchengshanense was 157,169 bp in length. It exhibited a typical quadripartite structure, including a large single-copy (LSC) region of 88,520 bp, a small single-copy (SSC) region of 17,075 bp and 2 inverted repeat regions (IRs) of 25,787 bp. The overall GC content was 37.78%, with 37.37%, 32.75%, and 43.18% for LSC, SSC and IR regions, respectively. The cp genome encoded 113 unique genes (79 protein-coding genes, 30 tRNA genes and 4 rRNA genes).
Phylogenetic analysis showed that E. jinchengshanense clustered with E. ilicifolium (NC_044897) with a moderate bootstrap value 85% (). The two Epimedium species were of morphological similarity of flowers and leaves, and they were distributed in the adjacent geographical areas, which might explain the close relationship between E. jinchengshanense and E. ilicifolium.
Author contribution
Y. X. and S. H. were involved in the conception and design; L. F. and X. H. contributed the sample collection; F. L. and C. W. performed the analysis and interpretation of the data; Y. X., L. F. and C. W. contributed the drafting of the paper; Y. X. and S. H. revised it critically for intellectual content. All authors were involved in the final approval of the version to be published. All authors agree to be accountable for all aspects of the work.
Ethics approval and consent to participate
The data collection of plants was carried out with permission of related institution, and complied with national or international guidelines and legislation.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MZ603799.1/ under the accession no. MZ603799. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA771137, SRR16347787, and SAMN22253056, respectively.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Guo MY, Pang XH, Xu YQ, Jiang WJ, Liao BS, Yu JS, Xu J, Song JY, Chen SL. 2022. Plastid genome data provide new insights into the phylogeny and evolution of the genus Epimedium. J Adv Res. 36:17–185.
- Jiang Y. 2020. Selection of medicinal germplasm of Epimedium L. [Master’s thesis]. Jiangxi University of Traditional Chinese Medicine; p. 37–42.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Liu SX, Liu LJ, Huang XF, Zhu YY, Xu YQ. 2017. A taxonomic revision of three Chinese spurless species of genus Epimedium L. (Berberidaceae). PK. 78:23–36.
- Stearn WT. 2002. The genus Epimedium and other herbaceous Berberidaceae. Portland: Timber Press; p. 27–134.
- Stéphane G, Francois DJ, Vincent L, Maria A, Wim H, Olivier G. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes . Nucleic Acids Res. 45(W1):W6–W11.
- Xu YQ, Jiang Y, Huang H, Li RE, Li FQ, Liu Y, Huang XF. 2020a. Taxonomic study of Epimedium L.: status, issues and prospect. Guihaia. 40(5):601–617.
- Xu YQ, Huang H, He YM, Li RQ, Huang XF. 2020b. Continuous petal variation in Epimedium tianmenshanense (Berberidaceae), a species endemic to western Hunan, China. Nord J Bot. 37(8):1–7.
- Xu YQ, Liu LJ, Liu SX, He YM, Li RQ, Ge F. 2019. The taxonomic relevance of flower colour for Epimedium (Berberidaceae), with morphological and nomenclatural notes for five species from China. PK. 118:33–64.
- Ying TS, Boufford DE, Brach AR. 2011. Epimedium L. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 19. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press; p. 787–799.
- Zhang YJ, Dang HS, Li JQ, Wang Y. 2014. The Epimedium wushanense (Berberidaceae) species complex, with one new species from Sichuan, China. Phytotaxa. 172(1):39–45.