Abstract
Pheidole nodus (Smith, 1874) belongs to a famously hyperdiverse and ecologically dominant ant genus. The mitochondrial genome of P. nodus is 15,579 bp in length, and the overall base composition is 78.6% AT. It includes 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNAs, and a control region. Phylogenetic trees show that P. nodus is more closely related to Wasmannia than to Atta. These sequence data will play an important role in the investigation of the phylogenetic relationships and taxonomy of the group Attini.
Pheidole, diet generalists of approximately 1200 known ant species (Bolton Citation2020), are found in most temperate and tropical biomes and on every continent except Antarctica and are particularly dominant in tropical habitats (Economo et al. Citation2015). Despite being the largest of all genera of plants and animals (Wilson Citation2003), Pheidole currently lacks a completely sequenced mitochondrial genome. Based on nuclear gene fragments, Pheidole is within the Attini group with Allomerus, Anisopheidole, Blepharidatta, Cephalotes, Diaphoromyrma, Lachnomyrmex, Lenomyrmex, Machomyrma, Ochetomyrmex, Procryptocerus, Tranopelta and Wasmannia, as well as all the fungus-growing ants (e.g. Atta) (Ward et al. Citation2015). However, based on the complete mitochondrial genome, the Attini group is polyphyletic because Wasmannia was shown to be closely related to the Solenopsidini group with strong support (Park et al. Citation2020; Yin et al. Citation2022). Pheidole nodus is widely distributed in eastern Asia, often occurring from open lands to relatively developed forests, and nests in the soil. Here, to better understand the phylogenetic relationship, we present the complete mitochondrial genome of Pheidole nodus as the first mitogenome of the genus Pheidole.
The specimens of Pheidole nodus workers were collected from a well-established colony in Nanchong City (30°49′25.30ʺN, 106°3′49.87ʺE), China, in October 2020. All protocols in the sample collection were reviewed and approved by the Research Ethics Committee of China West Normal University (Approval reference: CWNU2020D002). These specimens were preserved at −80 °C at the Key Laboratory of Southwest China Wildlife Resources Conservation, China West Normal University (www.cwnu.edu.cn; contact person: Yi LUO, [email protected]), after morphological identification under voucher number NCPN202010. Total genomic DNA was extracted and sequenced with the Illumina Novaseq sequencing platform with 150 bp paired-end reads by Shanghai Personal Biotechnology Co. Ltd, China. The genome de novo assembly was carried out with the software A5-miseq pipeline (Coil et al. Citation2015) and SPAdes V.3.14.1 (Bankevich et al. Citation2012). The MITOS Web Server (Bernt et al. Citation2013) was used for mitogenome annotation. The complete mitochondrial genome sequence information was deposited in GenBank under accession number MW429351.
The mitochondrial genome of P. nodus is 15,579 bp long and consists of 13 protein-coding genes (PCGs), 2 rRNAs, 22 tRNAs, and a control region. Its GC ratio is 21.4%. All PCGs used ATN (five ATG, four ATT and four ATA) as the start codon and TAA or TAG as the stop codon. The tRNA sizes range from 53 to 71 bp and are similar to those of other ants (approximately 54–90 bp) (Idogawa, Citation2021). Four PCGs (ND5, ND4, ND4L, and ND1) and ten tRNAS (tRNA-Val, Gln, Cys, Cys, Phe, His, Leu, Pro, and rRNA-Leu, Ser) are encoded by the majority strand (J-strand), while the others are located on the minority strand (N-strand). The gene order of P. nodus has two rearrangements compared to those of most Myrmicinae species: inversions between trnS1 and ND5 (Myrmicinae, trnE- trnF; P. nodus, trnF- trnE) and ND and ND6 (Myrmicinae, trnT- trnP; P. nodus, trnP-trnT). (Babbucci et al. Citation2014; Vieira and Prosdocimi Citation2019).
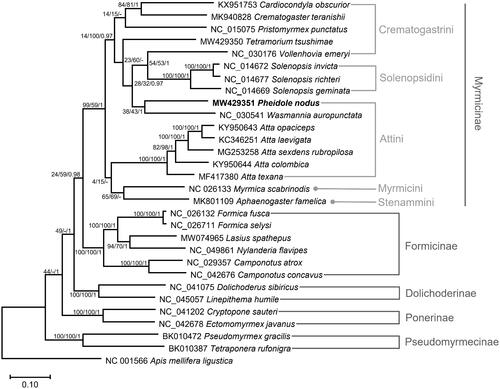
Thirteen PCGs and two rRNA genes from 29 ants, including P. nodus and an outgroup species, Apis mellifera ligustica, were aligned using MAFFT 7.450 (Katoh and Standley Citation2013) and concatenated for phylogenetic purposes in PhyloSuite v1.2.2 (Zhang et al. Citation2020). The GTR + F + I model was determined as a best-fit model by ModelFinder (Kalyaanamoorthy et al. Citation2017). Bootstrapped maximum likelihood, neighbor joining, and Bayesian inference trees were constructed using MEGA X (Kumar et al. Citation2018) and MrBayes 3.2.6 (Ronquist et al. Citation2012). The phylogenetic trees showed that Crematogastrini and Attini are polyphyletic with low support values (), which is congruent with the result based on the complete mitochondrial genome (Park et al. Citation2020; Yin et al. Citation2022) but disagrees with that based on nuclear gene fragments (Ward et al. Citation2015). Furthermore, Wasmannia showed a closer relationship to Pheidole than to Atta, which will aid our understanding of the phylogenetic relationship and taxonomy of the group Attini..
Figure 1. Maximum likelihood phylogenetic tree based on the concatenated PCGs and 2 rTNA genes of 30 Hymenoptera species (29 ants and one bee). Maximum likelihood and Bayesian inference phylogenetic trees were topologically identical. The numbers at the nodes indicate bootstrap support values of maximum likelihood and neighbor-joining trees and posterior probabilities of the Bayesian inference tree.
Author contributions
All authors contributed to the study conception and design. Analysis and interpretation of the data were performed by Yu Sang, Ru-Yi Yin and Yi Luo. The first draft of the manuscript was written by Yu Sang and Ru-Yi Yin. Yi Luo and Zhao-Min Zhou revised it critically for intellectual content, and all authors commented on previous versions of the manuscript. All authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW429351. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA772614, SRR16529670 and SAMN22402153, respectively.
Additional information
Funding
References
- Babbucci M, Basso A, Scupola A, Patarnello T, Negrisolo E. 2014. Is it an ant or a butterfly? Convergent evolution in the mitochondrial gene order of Hymenoptera and Lepidoptera. Genome Biol Evol. 6(12):3326–3343.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bolton B. 2020. AntCat. An online catalog of the ants of the world. [accessed 2021 Sep 11]. https://antcat.org.
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589.
- Economo EP, Klimov P, Sarnat EM, Guénard B, Weiser MD, Lecroq B, Knowles LL. 2015. Global phylogenetic structure of the hyperdiverse ant genus Pheidole reveals the repeated evolution of macroecological patterns. P Roy Soc B-Biol Sci. 282(1798):20141416.
- Idogawa N, Lee CC, Yang CCS, Dobata S. 2021. The complete mitochondrial genome of a parthenogenetic ant Monomorium triviale (Hymenoptera: Formicidae)). Mitochondr DNA B Resour. 6(10):2793–2795.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Park J, Xi H, Park J. 2020. The complete mitochondrial genome of Aphaenogaster famelica (Smith, 1874) (Hymenoptera: Formicidae). Mitochondr DNA B Resour. 5(1):492–494.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Vieira GA, Prosdocimi F. 2019. Accessible molecular phylogenomics at no cost: obtaining 14 new mitogenomes for the ant subfamily Pseudomyrmecinae from public data. PeerJ. 7:e6271.
- Ward PS, Brady SG, Fisher BL, Schultz TR. 2015. The evolution of Myrmicine ants: phylogeny and biogeography of a Hyperdiverse ant clade (Hymenoptera: Formicidae). Syst Entomol. 40(1):61–81.
- Wilson EO. 2003. Pheidole in the new world: a dominant, hyperdiverse ant genus. Cambridge, MA: Harvard University Press.
- Yin RY, Luo Y, Zhou ZM. 2022. The complete mitochondrial genome of Tetramorium tsushimae (Emery, 1925) (Hymenoptera: Formicidae). Mitochondr DNA B Resour. 7(1):40–42.
- Zhang D, Gao FL, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
