Abstract
Pithecellobium clypearia (Jack) Benth. 1844 belongs to the genus Pithecellobium in the family Fabaceae. The complete chloroplast genome of P. clypearia was sequenced and analyzed by Illumina sequencing in this study. The full length of the complete chloroplast genome is 176,770 bp, containing a pair of inverted repeat regions of 39,693 bp (IRa and IRb) separated by a large single-copy (LSC) region of 92,500 bp and a small single-copy (SSC) region of 4,884 bp. The P. clypearia chloroplast genome encodes 137 genes, comprising 92 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Phylogenetic analysis based on complete chloroplast genomes revealed that P. clypearia is closely related to Archidendron lucyi and Pithecellobium flexicaule. This study provides useful resources for further study and development of this species.
Pithecellobium clypearia (Jack) Benth. 1844 (homotypic synonym: Archidendron clypearia (Jack) I.C.Nielsen), is a species of the genus Pithecellobium in the family Fabaceae. It is widely distributed in southern China, such as Fujian, Guangdong, Guangxi, and Hainan provinces (Wu and Raven Citation2010). P. clypearia contains various compounds, including flavonoids, lignans, polyphenols, terpenoids, and steroids (Kang et al. Citation2014; Wang et al. Citation2020). It has been used to treat upper respiratory tract infections and gastroenteritis in traditional Chinese medicine (Thao et al. Citation2016).
The taxonomic status and phylogenetics position of P. clypearia remain controversial, some botanists thought it belongs to the genus Pithecellobium (Hooker Citation1844), while others thought it belongs to the genus Archidendron (Wu and Raven Citation2010). In recent years, comparative analysis of the complete chloroplast genome sequences has been used as an effective tool for studying plant phylogeny and origin (Liu et al. Citation2019; Chen et al. Citation2021). Here, in order to decipher the taxonomic status and phylogeny of P. clypearia, we analyzed its complete chloroplast genome and presented a phylogenetic analysis.
P. clypearia used for chloroplast genome sequencing was cultivated in the Biological Garden of Zhaoqing University (N23^6′, E112^30′, Zhaoqing, China). The voucher specimen (no. BGCLSZU003) was deposited in the Herbarium of Zhaoqing University (https://www.zqu.edu.cn/, Yinghua Wang, [email protected]). The total genomic DNA was extracted from fresh leaves of P. clypearia using the Plant Genomic DNA Kit (Tiangen, Beijing, China). Then, it was sequenced using the Illumina NovaSeq platform (Illumina, San Diego, CA). The raw data were mapped to plastome of Inga leiocalycina (GenBank accession number: KT428296.1), using Bowtie2 v2.2.4 (Langmead and Salzberg Citation2012) to exclude reads of nuclear and mitochondrial origins.
The chloroplast genome of P. clypearia was reconstructed with SPAdes 3.10.1 through the de novo assembly method (Bankevich et al. Citation2012), and de novo assembled chloroplast contigs were concatenated into larger contigs using Sequencher 5.3.2 (Gene Codes Inc., Ann Arbor, MI). Annotation of the chloroplast genome was generated by CpGAVAS (Liu et al. Citation2012) and a circular representation was drawn using the online tool OGDRAW (Lohse et al. Citation2007). The complete plastome sequence has been submitted to GenBank with the accession number of MW309810. The raw data have been uploaded to the NCBI Sequence Read Archive (SRA) database under the accession number SRR15400572.
The length of chloroplast genome sequence of P. clypearia is 176,770 bp, including two inverted repeat regions (IRa and IRb, each 39,693 bp) separated by a large single copy LSC (92,500 bp) region and a small single copy SSC (4,884 bp) region. The GC content of the overall chloroplast genome, IR regions, LSC, and SSC are 35.20%, 38.55%, 32.63%, and 29.30%, respectively. The chloroplast genome has 137 genes in total, including 92 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Among these identified genes, 44 genes are involved in photosynthesis, and 58 genes are involved in self replication.
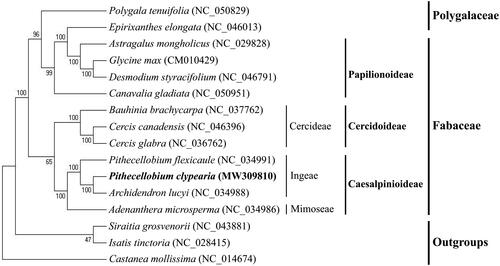
To analyze the phylogenetic position of P. clypearia, its plastome and the complete chloroplast genomes of 15 species from Fabaceae and its sister families published on NCBI GenBank were aligned using MAFFT v7.427 (Katoh et al. Citation2005). The gaps in the alignment were removed using the program trimAl with ‘-nogaps’ v 1.4 (Capella-Gutierrez et al. Citation2009). A maximum-likelihood (ML; 1000 bootstrap replicates) phylogenetic tree was constructed using MEGA v7.0 (Kumar et al. Citation2016), following the Kimura 2-parameter model of nucleotide substitution model. Siraitia grosvenorii, Isatis tinctoria, and Castanea mollissima were set as outgroups. The results showed that P. clypearia is closely related to Pithecellobium flexicaule and Archidendron lucyi in the tribe Ingeae (). The Leguminosae is the third largest angiosperm family in terms of species numbers, with close to 770 genera and over 19,500 species (LPWG Citation2013). In 2017, the Legume Phylogeny Working Group (LPWG) constructed a new phylogenetic tree of the family Leguminosae using an extensive sampling of plastomes. Based on the phylogenetic tree and taxonomic characters, LPWG proposed a new six-subfamily classification system (LPWG Citation2017). Wang et al. reconstructed a phylogenetic framework with strong statistical support, it was useful for future studies on legume classification, evolution, and diversification (Wang et al. Citation2017; Zhang et al. Citation2020). However, relationship between genera in the family Fabaceae has been difficult to resolve. This study showed P. clypearia is more closely related to A. lucyi than to P. flexicaule. Therefore, we suggest that P. clypearia belongs to the genus Archidendron.
Authors contributions
YW performed the experiments investigation, project administration, writing the original draft and data curation. GC prepared the resources, supervised the project, and made revisions to the manuscript.
Acknowledgements
Ethics statement: Ethical approval for the study was obtained from the Ethical Committee of Zhaoqing University.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that newly obtained at this study are available in the NCBI under accession number of MW309810 (https://www.ncbi.nlm.nih.gov/nuccore/MW309810). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA753227, SRR15400572, and SAMN20691823, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Chen G, Wang LG, Wang YH. 2021. Complete chloroplast genome sequence and phylogenetic analysis of Torenia fournieri. Mitochondrial DNA B Resour. 6(7):2004–2006.
- Hooker WJ. 1844. London journal of botany. Vol. 3. London (UK): Hippolyte Baillière; p. 209.
- Kang J, Liu C, Wang H, Li B, Li C, Chen R, Liu A. 2014. Studies on the bioactive flavonoids isolated from Pithecellobium clypearia Benth. Molecules. 19(4):4479–4490.
- Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33(2):511–518.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715.
- Liu XF, Zhu GF, Li DM, Wang XJ. 2019. Complete chloroplast genome sequence and phylogenetic analysis of Spathiphyllum 'Parrish'. PLOS One. 14(10):e0224038.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52(5–6):267–274.
- [LPWG] Legume Phylogeny Working Group. 2013. Legume phylogeny and classification in the 21st century: progress, prospects and lessons for other species-rich clades. Taxon. 62(2):217–248.
- [LPWG] Legume Phylogeny Working Group. 2017. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. Taxon. 66(1):44–47.
- Thao NP, Luyen B, Vinh LB, Lee JY, Kwon YI, Kim YH. 2016. Rat intestinal sucrase inhibited by minor constituents from the leaves and twigs of Archidendron clypearia (Jack.) Nielsen. Bioorg Med Chem Lett. 26(17):4272–4276.
- Wang YH, Qu XJ, Chen SY, Li DZ, Yi T. 2017. Plastomes of Mimosoideae: structural and size variation, sequence divergence, and phylogenetic implication. Tree Genet Genomes. 13:41.
- Wang YX, Duan ZK, Shi WY, Chang Y, Huang XX, Song SJ. 2020. A new dilignan from the twigs and leaves of Archidendron clypearia. J Asian Nat Prod Res. 23(6):1–6.
- Wu ZY, Raven P. 2010. Flora of China. Vol. 10 (Fabaceae). Beijing (China): Science Press.
- Zhang R, Wang Y-H, Jin J-J, Stull GW, Bruneau A, Cardoso D, De Queiroz LP, Moore MJ, Zhang S-D, Chen S-Y, et al. 2020. Exploration of plastid phylogenomic conflict yields new insights into the deep relationships of Leguminosae. Syst Biol. 69(4):613–622.