Abstract
Lonicera crassifolia is a prostrate or creeping, evergreen Lonicera species endemic to southwest China. Here, we reported the complete chloroplast (cp) genome of L. crassifolia (GenBank accession number: OK393707). The cp genome was 154,731 bp long, with a large single-copy region (LSC) of 88,619 bp and a small single-copy region (SSC) of 18,642 bp separated by a pair of inverted repeats (IRs) of 23,735 bp. It encodes 130 genes, including 85 protein-coding genes, 37 tRNA genes, and 8 ribosomal RNA genes. We also reconstructed the phylogeny of Lonicera using the maximum-likelihood (ML) method, including our data and previously reported cp genomes of related taxa. The phylogenetic analysis showed that L. crassifolia is a sister to the remaining Nintooa clade with strong bootstrap support.
The genus Lonicera L. is a major component of the family Caprifoliaceae, which comprises a large number of horticultural and economically important species (Ren et al. Citation2008; Jacobs et al. Citation2009). The genus includes 200 species of shrubs and climbers which are widely distributed in the Northern hemisphere (Rehder Citation1903, Citation1913; van Steenis Citation1946; Theis et al. Citation2008). Lonicera is well known for its taxonomic complexity, although has received extensive taxonomic evaluation and phylogenetic inference, the phylogenetic relationship among sections, subsections, and species is still unclear (Theis et al. Citation2008; Nakaji et al. Citation2015). Lonicera crassifolia Batalin 1892, which belongs to the section Nintooa, is a prostrate or creeping, evergreen shrub with axillary pink and yellow paired flowers, only occurs in streamsides, rocky cliffs, crevices of moist forest margins of southwest China (Hsu and Wang Citation1988). In order to better understand the phylogenetic relationship among the sections, subsections, and species of Lonicera, we assembled and characterized the complete chloroplast (cp) genome of L. crassifolia based on the Illumina pair-end sequencing data, and phylogenomic analysis of 20 Lonicera species was also presented.
Fresh leaves of L. crassifolia were collected from Wuguping, Xianfeng County, Hubei province (29.9860 N, 109.1126 E). A specimen was deposited at the Herbarium of Taizhou University (https://www.tzc.edu.cn/; collector: Zhong-Shuai Sun, [email protected]) under the voucher number LC1440-005. Total DNA was extracted from fresh leaves with a modified CTAB protocol (Doyle and Doyle Citation1987) and then sequenced using the Illumina NovaSeq platform (Illumina, San Diego, CA). The cp genome was assembled via NOVOPlasty (Dierckxsens et al. Citation2017), using the Lonicera japonica (NC026839) as the initial reference genome. The assembled cp genome was annotated using the online software GeSeq (Tillich et al. Citation2017) by comparing the sequences with the cp genome of L. japonica. Geneious R11 (Biomatters Ltd., Auckland, New Zealand) was used for inspecting the cp genome structure. The accurate annotated complete cp genome was submitted to GenBank with accession number OK393707.
The complete cp genome sequence of L. crassifolia is 154,731 bp in length, which has a characteristic quadripartite structure with a large single-copy (LSC) region of 88,619 bp, a small single-copy (SSC) region of 18,642 bp, and a pair of inverted repeats (IRs) of 23,735 bp. The overall GC contents of the total length, LSC, SSC, and IR regions were 38.6%, 37.0%, 33.4%, and 43.4%, respectively. The genome contained a total of 130 functional genes, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes.
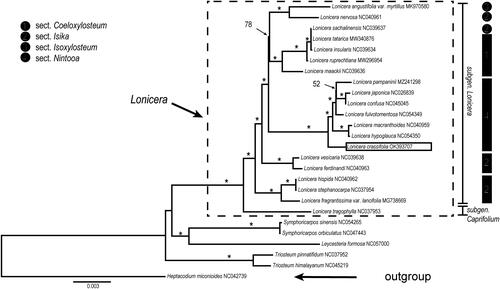
Species phylogeny intra Lonicera and the phylogenetic position of L. crassifolia were evaluated based on protein-coding regions (CDS) of the cp genome. 84 shared CDS were extracted from a total of 26 complete cp genomes from Caprifoliaceae and aligned by MAFFT (Katoh et al. Citation2005). Heptacodium miconioides (NC042739) was used as an outgroup. We reconstructed a phylogeny employing the GTR + G model and 1000 bootstrap replicates under the maximum-likelihood (ML) inference in RAxML-HPC v.8.2.10 on the CIPRES cluster (Miller et al. Citation2010). A robust phylogeny of Lonicera was obtained based on the CDS data, and the genus was resolved as a monophyletic clade consisting of two well-supported clades, subgen. Lonicera and subgen. Caprifolium (). Our result supports the classification of the two subgenera in Lonicera proposed by Rehder (Citation1903, Citation1913) and Hsu and Wang (Citation1988) and coincides with previous molecular phylogenetic studies (Theis et al. Citation2008; Nakaji et al. Citation2015; Wu et al. Citation2021). Among the four sections of subgen. Lonicera proposed by Rehder (Citation1903, Citation1913) and Hsu and Wang (Citation1988), sect. Nintooa was supported as monophyly, and L. crassifolia was placed at the base of the Nintooa clade with strong bootstrap support ().
Ethical approval
The material involved in the article does not involve ethical conflicts. This study was permitted by the Zhejiang Provincial Key Laboratory of Plant Evolutionary Ecology and Conservation, Taizhou University, China. All collection and sequencing works were strictly executed under local legislation and related laboratory regulations to protect wild resources.
Authors’ contributions
The article was designed and conceived by Chao Chen and Zhong-Shuai Sun; Chao Chen and Da-Hao Qu assembled and annotated the cp genome; Fang-Quan Shan collected and identified the plant material; Zhong-Shuai Sun contributed significantly to phylogenetic analysis and manuscript preparation; Ze-Xin Jin was involved in the interpretation of the data and revised the manuscript critically for intellectual content. All authors approved the final version to be published and agreed to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. OK393707. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA768984, SRR16235359 and SAMN22073044, respectively.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hsu PS, Wang HJ. 1988. Lonicera. In: Hsu PS, editor. Flora Reipublicae Popularis Sinicae. Vol. 72. Beijing (China): Science Press; p. 143–259.
- Jacobs B, Lens F, Smets E. 2009. Evolution of fruit and seed characters in the Diervilla and Lonicera clades (Caprifoliaceae, Dipsacales). Ann Bot. 104(2):253–276.
- Katoh K, Kuma KI, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33(2):511–518.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Comp Env. 14:1–8.
- Nakaji M, Tanaka N, Sugawara T. 2015. A molecular phylogenetic study of Lonicera L. (Caprifoliaceae) in Japan based on chloroplast DNA sequences. Acta Phytotaxon Geobot. 66(3):137–151.
- Rehder A. 1903. Synopsis of the genus Lonicera. Missouri Bot Gard AnnRep. 1903:27–232.
- Rehder A. 1913. Caprifoliaceae. In Sargent CS, editor. Plantae Wilsonianae: an enumeration of the woody plants collected in western China for the Arnold Arboretum of Harvard University during the years 1907, 1908, and 1910. Cambridge (MA): Harvard University Press; p. 106–144.
- Ren MT, Chen J, Song Y, Sheng LS, Li P, Qi LW. 2008. Identification and quantification of 32 bioactive compounds in Lonicera species by high performance liquid chromatography coupled with time-of-flight mass spectrometry. J Pharm Biomed Anal. 48(5):1351–1360.
- Theis N, Donoghue MJ, Li JH. 2008. Phylogenetics of the Caprifolieae and Lonicera (Dipsacales) based on nuclear and chloroplast DNA sequences. Syst Bot. 33(4):776–783.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- van Steenis CGGJ. 1946. Preliminary revision of the genus Lonicera in Malaysia. J Arnold Arboretum. 27:442–452.
- Wu YM, Ma D, Zhu YX, Liu B, Mu XY. 2021. Characterization of the complete chloroplast genome of a subalpine deciduous shrub Lonicera angustifolia var. myrtillus (Caprifoliaceae). Mitochondrial DNA B Resour. 6(7):1796–1798.