Abstract
Machilus salicina Hance, 1885 is a species of flowering plant in the family Lauraceae and is mainly found at low altitudes in southern China. In this study, we assembled and annotated the complete chloroplast genome of M. salicina for the first time. We analyzed the general features of M. salicina and constructed a phylogenetic tree based on 15 Lauraceae species. The chloroplast genome of M. salicina had a total length of 153,943 bp. The length of a large single copy region, a small single copy region, and two inverted repeat regions were 93,689 bp, 20,070 bp, and 20,092bp, respectively. A total of 128 genes were detected, which included 84 protein-coding genes, 36 tRNAs and 8 rRNAs. The GC content of M. salicina complete chloroplast genome was 39.1%. The phylogenetic tree indicated that M. salicina was closely related to M. yunnanensis.
Machilus salicina Hance, 1885, is a species of flowering plant in the family Lauraceae and is mainly found at low altitudes in Guangdong, Guizhou, and Yunnan provinces of China. The leaves of M. salicina are dense and can be used as herbs to help reduce swelling and detoxify the body (Zhijun and Young Citation2003; Zuo et al. Citation2012; Chen et al. Citation2020). As the cost of next-generation sequencing technology has decreased, chloroplast genomes have increasingly been sequenced (Ge et al. Citation2019; Song et al. Citation2022). The chloroplast genomes of Machilus (M. yunnanensis, M. balansae, and M. robusta) have been reported in previous studies (Song et al. Citation2015; Wu et al. Citation2021). However, the chloroplast genome of M. salicina has yet to be reported. In this study, we assembled and annotated the complete chloroplast genome sequence of M. salicina for the first time. We analyzed the general features of M. salicina and constructed a phylogenetic tree based on 15 Lauraceae species. This study will contribute to species identification and ecological conservation.
Fresh leaves of M. salicina were sampled from Panlong District, Kunming City, Yunnan Province, China (24°23′N, 102°10′E). We conducted this research with the consent of the local government and the Kunming Institute of Botany, Chinese Academy of Sciences. The voucher samples and DNA were stored at Qingdao University of Science and Technology (Chao Shi, [email protected]) under the specimen code MC202114. Fresh leaves tissue with no obvious signs of disease were collected and preserved in silica gel. Complete genomic DNA was extracted from fresh leaves using modified CTAB (Porebski et al. Citation1997). Both the quantity and quality of the extracted DNA were assessed spectrophotometrically, while the integrity was assessed using 1% (w/v) agarose gel electrophoresis. Illumina TruSeq library preparation kits (Illumina, San Diego, CA, USA) were used to prepare DNA insert libraries of approximately 500 bp paired ends according to the manufacturer's protocol. These libraries were sequenced on the Illumina HiSeq 4000 platform in Novogene (Beijing, China) and generated raw data of 150 bp paired-end reads. About 4.3 Gb high quality, 2 × 150 bp paired-end raw reads were obtained and were used to assemble the complete chloroplast genome of M. salicina (Wang et al. Citation2018). We assembled the chloroplast genome of M. salicina using NOVOPlasty v4.3.1 (Dierckxsens et al. Citation2017) and annotated the chloroplast genome with GeSeq (Tillich et al. Citation2017). We used Sequin to manually correct codons and gene boundaries.
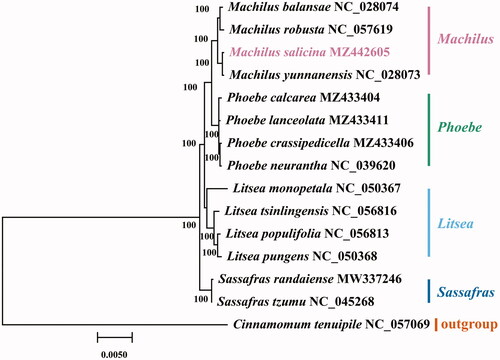
The chloroplast genome of M. salicina (GenBank accession MZ442605) presented a typical quadripartite structure (Wicke et al. Citation2011) with a total length of 153,943 bp. The large single copy (LSC) region was 93,689 bp in length, and the small single copy (SSC) region was 20,070 bp in length. Two inverted repeat (IR) regions were 20,092 bp in length. A total of 128 genes were found in the chloroplast genome of M. salicina, with a GC content of 39.1%. About 128 genes were comprised of 84 protein-coding genes, 36 tRNAs and 8 rRNAs. We performed a phylogenetic analysis of M. salicina to reveal its evolutionary relationships with other Lauraceae species. The chloroplast genome sequences of 14 Lauraceae species were obtained from NCBI. Cinnamomum tenuipile was used as an outgroup. The chloroplast genomes of the 14 Lauraceae and the newly assembled M. salicina were aligned using MAFFT v725 (Katoh and Standley Citation2013). GTR-GAMMA (GTR + G) model was selected by Modeltest (Posada and Crandall Citation1998) based on the Bayesian information content (BIC) criterion. The maximum likelihood (ML) method was used for 1000 bootstraps by MEGA-X software (Kumar et al. Citation2018). The phylogenetic tree showed that M. salicina closely was related to M. yunnanensis and Machilus was closely sister to Phoebe (). There were parallels between these phenomena and previous studies (Chen et al. Citation2017; Song et al. Citation2017).
Ethics statement
This research was conducted with the consent of the local government and the Kunming Institute of Botany, Chinese Academy of Sciences. The collection of plant materials is carried out in accordance with guidelines provided by the Kunming Institute of Botany, Chinese Academy of Sciences and Yunnan province regulations. Field studies have complied with local legislation, and the manuscript includes a statement of appropriate permissions granted and licenses from related agencies of Yunnan province.
Author contributions
Xiaoxuan Long: Conceptualization, Writing - Original Draft. Wenbo Shi: Data Curation, Software. Weicai Song: Methodology, Software. Weiqi Han: Data Curation. Guiwen Yang: Visualization, Data Curation. Shuo Wang: Data Curation, Resources, Writing - Reviewing and Editing. All authors have read and approved the final manuscript.
Acknowledgements
We are thankful to Beijing-based Novogene for their NGS service that was instrumental to the execution of the project. We are grateful to Chao Shi and Zimeng Chen for their guidance and assistance in the assembly of the chloroplast genome of Machilus salicina in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession MZ442605. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA786751, SRR17153751, and SAMN23720018, respectively.
Additional information
Funding
References
- Chen C, Zheng Y, Liu S, Zhong Y, Wu Y, Li J, Xu LA, Xu M. 2017. The complete chloroplast genome of Cinnamomum Camphora and its comparison with related Lauraceae species. PeerJ. 2017(9):1–18.
- Chen YC, Li Z, Zhao YX, Gao M, Wang JY, Liu KW, Wang X, Wu LW, Jiao YL, Xu ZL, et al. 2020. The Litsea genome and the evolution of the Laurel family. Nat Commun. 11(1):1–14.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):1–9.
- Ge Y, Dong X, Wu B, Wang N, Chen D, Chen H, Zou M, Xu Z, Tan L, Zhan R. 2019. Evolutionary analysis of six chloroplast genomes from three Persea Americana ecological races: insights into sequence divergences and phylogenetic relationships. PLoS ONE. 14(9):e0221827–17.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 15(1):8–15.
- Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics. 14(9):817–818.
- Song W, Ji C, Chen Z, Cai H, Wu X, Shi C, Wang S. 2022. Comparative analysis the complete chloroplast genomes of nine Musa species: genomic features, comparative analysis, and phylogenetic implications. Front Plant Sci. 13(February):1–15.
- Song Y, Dong W, Liu B, Xu C, Yao X, Gao J, Corlett RT. 2015. Comparative analysis of complete chloroplast genome sequences of two tropical trees Machilus Yunnanensis and Machilus Balansae in the family Lauraceae. Front Plant Sci. 6(Aug):662–668.
- Song Y, Yao X, Tan Y, Gan Y, Yang J, Corlett RT. 2017. Comparative analysis of complete chloroplast genome sequences of two subtropical trees, Phoebe Sheareri and Phoebe Omeiensis (Lauraceae). Tree Genet Genomes. 13(6):120.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–11.
- Wang X, Cheng F, Rohlsen D, Bi C, Wang C, Xu Y, Wei S, Ye Q, Yin T, Ye N. 2018. Organellar genome assembly methods and comparative analysis of horticultural plants. Hortic Res. 5(1):3–13.
- Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76(3-5):273–297.
- Wu Y, Zheng Y, Liu X, Dai X, Li S, Xu H. 2021. The complete chloroplast genome sequence of Machilus Robusta W. W. Smith (Lauraceae) from Jiangxi Province, China. Mitochondrial DNA Part B. 6(7):1890–1892.
- Zhijun W, Young SS. 2003. Differences in bird diversity between two swidden agricultural sites in mountainous terrain, Xishuangbanna, Yunnan, China. Biol Conserv. 110(2):231–243.
- Zuo GY, Zhang XJ, Yang CX, Han J, Wang GC, Bian ZQ. 2012. Evaluation of traditional Chinese medicinal plants for anti-MRSA activity with reference to the treatment record of infectious diseases. Molecules. 17(3):2955–2967.