Abstract
We sequenced and annotated the complete mitochondrial genome of Achroia grisella (Fabricius, 1794), which was 15,368 bp, encoding 13 protein-coding genes (PCGs), 22 tRNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and a control region; having a base composition of A (38.7%), C (12.3%), G (7.5%), T (41.5%), G + C (19.8%), and A + T (71.2%); except for the ND1, which was initiated by the GAT codon, the other 12 PCGs were initiated by the ATN (ATT, ATA, and ATG) codon. The nine PCGs terminated with the typical TAA stop codon, whereas the remaining four genes had incomplete stop codons, which were single T. In addition, the phylogenetic relationships based on nucleotide sequences of 13 PCGs using Bayesian inference (BI) and maximum likelihood (ML) methods showed a close relationship between Achroia grisella and Galleria mellonella.
The small wax moth, Achroia grisella Fabricius, belongs to the Lepidoptera family Pyralidae genus Galleria Fabricius insects, and both Achroia grisella larvae and Galleria mellonella Linne larvae are collectively referred to as nests pest (Wen Citation2008). The damage of Galleria mellonella to honeybees commonly occurs in bee-keeping areas of South China. A. grisella lives at the bottom of the hive and feeds on wax chips dropped from the bottom of the hive, which can also destroy the unpreserved nest spleen. (Seeley Citation2009; Zhang et al. Citation2019; Mahgoub et al. Citation2020). The larva of the great wax moth feeds on the nest spleen of bees and causes a large number of capped pupae to die or the bee colony to give up the nest, which is seriously harmful (Kwadha et al. Citation2017). Therefore, less attention has been paid to the small wax moth, and there is some confusion in morphological identification between A. grisella and G. mellonella, which are not easy to identify (Zhang et al. Citation2019). In this study, we focused on the mitochondrial genome of A. grisella in an attempt to analyze the phylogenetic relationship and evolutionary relationship of A. grisella, which has important biological significance for the species identification and genetic background of Achroia grisella from the molecular biological level.
The specimens used in this study were collected from Yongai Village, Shangtang Town, Kaili City, Guizhou Province (Miaojiang bee breeding base, Huangping County) (107.69E, 26.91 N) and were deposited in the Guizhou Provincial key laboratory for rare animal and economic insects of mountainous region (Ms. Liu, [email protected]) (Certificating number: GY2021090101). Among them, the common method to collect insect samples is to immerse the insects in alcohol immediately after they are captured as a sample for DNA extraction. This does not torture insects and is not unethical.
A sample’s total genomic DNA was extracted from each individual using the TIANGEN Genomic DNA Extraction Kit (TIANGEN Co., Beijing, China) according to the manufacturer's protocol, sequencing libraries with an average length of 350 bp were generated using the TruSeq Nano DNA HT Sample Preparation Kit (Illumina USA), and then the libraries were sequenced on the Illumina Novaseq 6000 platform. After quality checking of the obtained sequences, raw sequence reads were edited using the NGS QC toolkit (Patel and Jain Citation2012), clean data were de novo assembled by Mitoz v. 2.3 software (Meng et al. Citation2019). Finally, annotation of the mitochondrial genomes was performed using Geneious R10 (Biomatters, Auckland, New Zealand), with BLAST against and related species from GenBank as the reference (Altschul et al. Citation1990; Kearse et al. Citation2012), and submitted to GenBank (accession number: OM203125) (https://www.ncbi.nlm.nih.gov/.) and SRA (accession number: PRJNA792436) (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA792436).
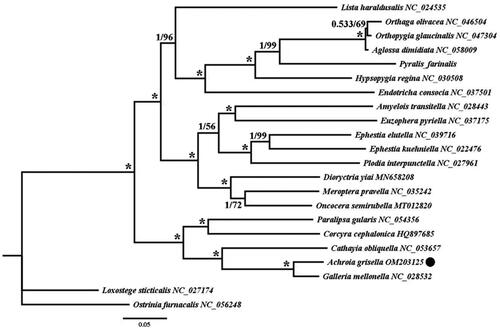
Twenty-two mitogenomic sequences (downloaded from GenBank) were used in the phylogenetic analysis, and 13 coding protein genes of all 21 insect species and Achroia grisella were extracted using Geneious R10, with Loxostege sticticalis and Ostrinia furnacalis as outgroups. We then used Clustal X 1.83 (Thompson et al. Citation1997) and SequenceMatrix 1.8 using default settings (Vaidya et al. Citation2011), and 13 protein-coding genes were concatenated as a dataset. Nucleotide matrices were used for phylogenetic analysis using two approaches: Bayesian inference (Ronquist et al. Citation2012) and Maximum likelihood (Nguyen et al. Citation2015). The best-fit models (GTR + I + G and GTR + F + I + G4) for the nucleotide sequence were selected using the Akaike Information Criterion (AIC) in jModeltest 2.1.10 and IQ-TREE 1.6.2, respectively (Darriba et al. Citation2012; Nguyen et al. Citation2015). The resulting phylogenetic tree is edited and visualized in FigTree 1.4.0 (Mousavi et al. Citation2014) ().
Figure 1. The phylogenetic trees of Achroia grisella were inferred from the amino acid sequences of the 13 PCGs in mitogenome of 22 species from using Bayesian inferences (BI) and maximum likelihood (ML) methods. Numbers on branches indicate BI posterior probability (PP, left) and ML bootstrap support (BS, right), respectively. An asterisk denotes PP =1 and BS = 100. The black dot indicates mitogenome of Achroia grisella newly determined in this study.
The complete mitochondrial genome sequence of A. grisella (GenBank: OM203125) is 15,368 bp, with base compositions of A = 38.7%, C = 12.3%, G = 7.5%, and T = 41.5%. Our sequences are similar to those of other metazoans, which contain 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a non-coding AT-rich region. Except for the ND1, which is initiated by a particular GAT codon, the other 12 PCGs are all initiated by ATN (ATT, ATA, and ATG) codons. Of these, three PCGs (COX3, ND4L, and ATP6) started with ATG, seven PCGs (COX1, COX2, ND2, ND3, ND4, ND5, and ATP8) started with ATT, one PCG (CYTB) started with ATA, and one PCG (ND6) started with ATC; in addition, nine PCGs (ND2, ND3, ATP8, ATP6, ND4L, ND5, ND6, COX3, and CYTB) terminated with TAA codons and four PCGs (ND1, ND4, COX1, and COX2) had incomplete stop codons, which were single T.
Phylogenetic relationship analysis () showed that A. grisella belongs to the genus Galleriinae and has the highest homology and closest genetic relationship with Galleria mellonella, which is consistent with the results of phylogenetic analysis of the small wax moth based on the cytochrome c oxidase subunit I gene by Zhang et al. (Citation2019). In addition, it was found that A. grisella was clustered with other Pyralidae insects. Therefore, according to the analysis, we could further reflect the evolutionary relationship of A. grisella and clarified that the complete mitochondrial genome sequence of A. grisella was the evolution of the Pyralidae, thus providing a theoretical basis for exploring the genetic diversity and phylogenetic study of the main enemy of the colony.
Authors’ contributions
Yangyang Liu, analyzed the data, performed the experiments, involved in certain tools for analysis, prepared figure, drafted of the paper, and approved the final draft.
Guoyong Li, collected and analyzed data, consulted the relevant literature, and uploaded the analysis data.
Jiankun Long, contributed reagents/materials, involved in conception and design of the work, and approved and published the final draft.
All authors agree to be accountable for all aspects of the work.
Disclosure statement
The authors reported no potential conflicts of interest.
Data availability statement
The data that support the findings of this study are openly available at NCBI database. The GenBank Accession No. (reference number) is OM203125 (https://www.ncbi.nlm.nih.gov/.). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA792436, SRS11402858, and SAMN24425024, respectively, and all of the accession numbers are activated.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper S, Markowitz A, Duran S, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kwadha CA, Ong’Amo GO, Ndegwa PN, Raina SK, Fombong AT. 2017. The biology and control of the greater wax moth, Galleria mellonella. Insects. 8(2):61.
- Mahgoub MO, Wei HL, Omar DB, Naim A. 2020. Life table and demographic parameters of the lesser wax moth, Achroia grisella, reared on natural honey bee wax. WJAR. 8(1):12–15.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. Mitoz: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Mousavi SA, Österman J, Wahlberg N, Nesme X, Lavire C, Vial L, Paulin L, de Lajudie P, Lindström K. 2014. Phylogeny of the rhizobium-allorhizobium-agrobacterium clade supports the delineation of neorhizobium gen. nov. Syst Appl Microbiol. 37(3):208–215.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7(2):e30619.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Seeley TD. 2009. The wisdom of the hive: the social physiology of honey bee colonies. Cambridge: Harvard University Press.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25(24):173–216.
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 27(2):171–180.
- Wen B. 2008. Achroia grisella Fabricius and Galleria mellonella Linne. Apicult China. 59(7):29–29.
- Zhang YH, Zhu F, Tang FF, Shao YL, Yang S, Bai XR. 2019. Phylogenetic analysis of Achroia grisella Fabricius based on the mitochondrial cytochrome c oxidase subunit I gene. Chin J Appl Entomol. 56(1):107–113.
