Abstract
The complete plastid genome of Scutellaria microviolacea C. Y. Wu was firstly reported. The full length of the plastid genome was 152,092 bp and comprised of a large single-copy (LSC) region of 84,090 bp, a small single-copy (SSC) region of 17,534 bp, and two inverted repeat regions (IRs) of 25,234 bp. A total of 131 genes were encoded, including 87 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The overall GC content of the S. microviolacea plastid genome was 38.3%. Further phylogenetic analysis based on 18 accessions inferred that the genus Scutellaria can be divided into two clades, and S. microviolacea is evolutionarily close to Scutellaria tsinyunensis. Our study provided essential genetic resources for further studies on the evolution and genetic diversity of the genus Scutellaria and its related taxa.
The genus Scutellaria belongs to the subfamily of Scutellarioideae (Lamiaceae), which contains about 360 species and is widely distributed in East Asia, Europe, and the United States (Paton Citation1990; Paton et al. Citation2016). Many species of Scutellaria possess a wide range of pharmacological actions, such as antitumor, antioxidant, anticonvulsant, antibacterial, antiviral, anti-angiogenesis, and hepatoprotective activities (Shang et al. Citation2010). The infrageneric classifications and species delimitation of this genus remain to be controversial (Shang et al. Citation2010; Zhao et al. Citation2021). S. microviolacea C. Y. Wu Citation1977 is an endemic species from Yunnan Province of China (Wu Citation1977). In the present study, we assembled and characterized the first complete plastid genome of S. microviolacea and performed the phylogenetic analysis with other 13 congeneric species.
The sample of S. microviolacea was collected from Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences. Total genomic DNA was extracted from leaf tissue using the CTAB method (Doyle and Doyle Citation1987). The voucher specimen (Voucher number: 08CS573, Collected By Ting Zhang, [email protected]) was deposited in the Herbarium of the Kunming Institute of Botany, Chinese Academy of Sciences, Yunnan, China (KUN). Genome skimming sequencing was performed on the Illumina HiSeq X Ten platform (Illumina, San Diego, CA). Totally, 3.15 Gb high-quality clean data were generated. The GetOrganelle v1.7.0 (Jin et al. Citation2020) pipeline was used to assemble the complete plastid genome with clean sequencing data. The Bandage v 0.8.1 (Wick et al. Citation2015) was used to visualize the completeness of genome assembly. Finally, the PGA (Qu et al. Citation2019) was used to annotate all genes of the plastid genome with the whole plastid genome sequence of Scutellaria lateriflora (NC_034693) as reference. The newly annotated plastid genome was available in GenBank of NCBI (accession number: MZ954872.1).
The complete plastid genome of S. microviolacea was 152,092 bp in length and comprised of a pair of inverted repeats (IRs) of 25,234 bp each, a large single-copy (LSC) region of 84,090 bp, and a small single-copy (SSC) region of 17,534 bp. The overall GC content of this genome was 38.3% (IRs, 43.6%; LSC, 36.4%; SSC, 32.5%). Totally, the plastid genome encoded 131 genes, including 87 protein-coding genes, 36 tRNA genes, and 8 rRNA genes.
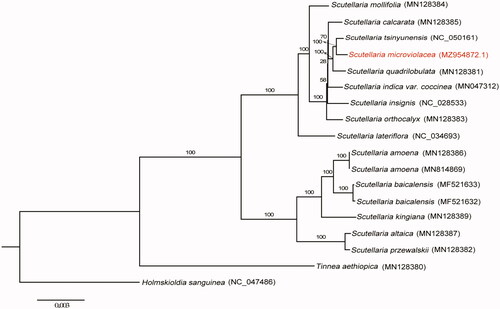
To clarify the phylogenetic position of S. microviolacea, 16 plastomes involving 14 species from the genus of Scutellaria were used to perform the maximum likelihood (ML) analysis (). Due to the successive sister groups’ relationship among Tinnea, Holmskioldia, and Scutellaria (Zhao et al. Citation2021), we chose Tinnea aethiopica, Holmskioldia sanguinea as outgroups. All plastid genomes were aligned by MAFFT (Katoh and Standley Citation2013) plugin in the software PhyloSuite (Zhang et al. Citation2020). The ML analysis was performed using RAxML (Stamatakis Citation2006) under the GTR + G model, and the rapid bootstrap was set to 1000 replicates. Based on our phylogeny, the genus Scutellaria was divided into two clades, which is consistent with previous studies despite different samplings being used (Safikhani et al. Citation2018; Zhao et al. Citation2021). The endemic species S. microviolacea is evolutionarilyclose to Scutellaria tsinyunensis. Given the species richness and complex taxonomy history in the genus Scutellaria, sufficient species sampling with genomic sequencing data is necessary to accurately authenticate the interspecific relationship of Scutellaria. The newly characterized complete plastid genome of S. microviolacea will provide essential resources for further studies on the evolution and genetic diversity of the genus Scutellaria and its related taxa.
Ethical approval
The sample collection was authorized by Professor Jun-Bo Yang (Germplasm Bank of Wild Species, Kunming Institute of Botany, CAS).
Author contributions
YC Wang drafted the manuscript; YC Wang and ZR Zhang performed the data analysis; LJ Yan designed this study and revised the manuscript critically for intellectual content; all authors contributed to the final approval of the version to be published and all agree to be accountable for all aspects of the work.
Acknowledgments
We are grateful to Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences for providing the sample of Scutellaria microviolacea.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ954872.1. The associated BioProject, Bio-Sample and SRA, numbers are PRJNA758007, SAMN20989313, and SRR15672851 respectively.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241–231.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Paton A. 1990. A global taxonomic investigation of Scutellaria L. and its allies (Labiatae). Kew Bull. 45(3):399–450.
- Paton A, Suddee S, Bongcheewin B. 2016. Two new species of Scutellaria (Lamiaceae) from Thailand and Burma. Kew Bull. 71(1):1–6.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(50):50–12.
- Safikhani K, Jamzad Z, Saeidi H. 2018. Phylogenetic relationships in Iranian Scutellaria (Lamiaceae) based on nuclear ribosomal ITS and chloroplast trnL-F DNA data. Plant Syst Evol. 304(9):1077–1089.
- Shang X, He X, He X, Li M, Zhang R, Fan P, Zhang Q, Jia Z. 2010. The genus Scutellaria an ethnopharmacological and phytochemical review. J Ethnopharmacol. 128(2):279–313.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Wu ZY. 1977. Flora Yunnanica Tomus 1. Beijing (China): Science Press.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
- Zhao F, Chen YP, Salmaki Y, Drew BT, Wilson TC, Scheen AC, Celep F, Bräuchler C, Bendiksby M, Wang Q, et al. 2021. An updated tribal classification of Lamiaceae based on plastome phylogenomics. BMC Biol. 19(1):2–27.