Abstract
Iris speculatrix is a rare and endangered plant first discovered in and native to Hong Kong. The whole chloroplast genome of I. speculatrix is 152,368 bp in length. It contained a large single copy region (82,003 bp), a small single copy region (17,941 bp), and two inverted repeats (26,212 bp). Phylogenetic analysis of 17 species of Iridaceae was conducted. I. speculatrix was found to be sister to a group of 12 Iris species, including I. setosa, I. lacteal, and I. uniflora. The sequenced chloroplast whole genome would be useful to understand the phylogeny and to conservation of I. speculatrix.
Content
Iris speculatrix Hance (Iridaceae) (Hance Citation1875), a herb with attractive purplish flower, was first discovered and published in Hong Kong and is a rare and precious plant of Hong Kong scheduled under the Forestry Regulations (Hu et al. Citation2003). Members of the genus Iris are well known for their attractive flowers and are widely applied in horticulture (Kang et al. Citation2020).
The sample of I. speculatrix was collected in Tai Tam, Hong Kong, in collaboration with Agriculture, Fisheries and Conservation Department (AFCD), The Government of the Hong Kong Special Administrative Region (HKSAR) as part of the Rare and Endangered Species project. The species is scheduled under protective legislation (and exact locality details are restricted). Hence, it is required by AFCD, The Government of the Hong Kong Special Administrative Region that the GPS should be kept confidential. The voucher specimens with collector number Rare and endangered plants (SW Shek, KW Lam, DTW Lau, and JYY Lau) 009 were deposited in the Hong Kong Herbarium, AFCD, HKSAR (https://www.herbarium.gov.hk/en/home/index.html, Dr. LAU Yuen Yung, Jenny, [email protected]) and the Shiu-Ying Hu Herbarium, School of Life Sciences, The Chinese University of Hong Kong (https://syhuherbarium.sls.cuhk.edu.hk/, David Tai Wai Lau, [email protected]).
Fresh leaves of Iris speculatrix (60 mg) was used for the extraction of total genomic DNA. The sample was extracted using i-genomic Plant DNA Extraction Mini Kit (iNtRON Biotechnology, Seongnam, Gyeonggi, Republic of Korea) The manufacturer’s protocol was used. The concentration of the extract was checked using NanoDrop Lite (Thermo Fisher Scientific, Massachusetts, USA). Then, 1% agarose gel electrophoresis was used to visualize and check DNA quality. The sample was sequenced using NovaSeq 6000 platform (Illumina Inc., San Diego, CA, USA) by Novogene Bioinformatics Technology Co., Ltd. (https://en.novogene.com/, Beijing, China). A 150 bp paired-end (PE) library was constructed by Illumina sequencing. CLC Assembly Cell package v5.1.1 (CLC Inc., Denmark) was used to trim poor-quality reads (Phred score < 33).
CLC de novo assembler in CLC Assembly Cell package and SOAPdenovo v3.23 were used to assemble reads into contigs under default parameters. Gapcloser module in SOAP package was used to close gaps. After that, the contigs were aligned to the reference genome Iris rossii (NC_056180.1), and assembled into complete chloroplast genome. Gene annotation was obtained from GeSeq platform with reference to complete chloroplast genomes of Iris domestica (NC_050833.1) and Iris gatesii (NC_024936.1). A few adjustments were made manually for protein-coding genes, start and stop codons. The annotated genome was deposited in GenBank with the accession number OK274247.
The complete chloroplast genome of I. speculatrix was 152,368 bp in length, containing a large single copy (LSC) region (82,003 bp), a small single copy region (17,941 bp), and two inverted repeats (26,212 bp). The GC content was 38.04%. The chloroplast genome contained 112 unique genes, including 79 protein-coding genes, 29 tRNA genes, and 4 rRNA genes.
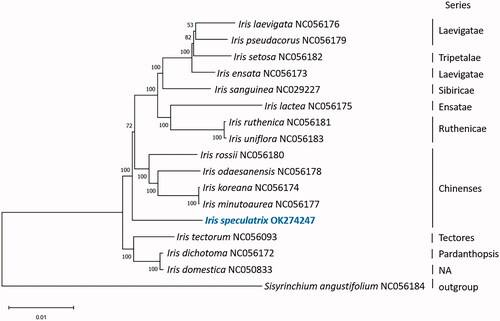
To investigate the taxonomic position of I. speculatrix, the chloroplast genome was aligned with 15 other species in the genus Iris, and the Sisyrinchium angustifolium (NC056184) within Iridaceae was used as an outgroup. The complete chloroplast genomes were aligned using MAFFT 7.48 (Katoh et al. Citation2019). A maximum likelihood (ML) tree was constructed using MEGA X (Kumar et al. Citation2018) based on the best fit model GTR + G and 1000 bootstrap replicates. The phylogenetic tree was labeled with species and series (). I. speculatrix was found to be sister to a group of 12 Iris species, including I. setosa, I. lactea and I. uniflora. In previous study of Iris phylogeny based on trnL-trnF and rps4 data, I. speculatrix was sister to a group of 17 Iris species including I. xiphium and I. latifolia, while the group was sister to another group of Iris containing I. setosa, I. lactea and I. uniflora (Tillie et al. Citation2000). From Tillie’s study, I. speculatrix was more distantly related to the Chinenses group of Iris including I. rossii. However, from our phylogenetic analysis, I. speculatrix was sister to a larger group of Iris, which included I. rossii and I. koreana from series Chinenses, I. setosa from the series Tripetalae. The species was more distantly related to I. tectorum from series Tectores and I. dichotoma from Pardanthopsis.
It was proposed that Iris from the series Chinenses should be classified to the new genus Zhaoanthus nom. provis. (Mavrodiev et al. Citation2014). Although it was suggested that I. speculatrix should be moved to Zhaoanthus based on morphological characters (Crespo et al. Citation2015; Wilson Citation2020), the treatment was not accepted by The World Checklist of Vascular Plants (Govaerts et al. Citation2021), where Zhaoanthus speculatrix was treated as a synonym of I. speculatrix.
In another study, I. speculatrix was also found to be outside of the Chinenses group (Guo and Wilson Citation2013). Tillie had suggested that I. speculatrix was highly sequence-divergent compared to other members of the group including I. xiphium and I. latifolia. Previous study had also suggested that I. speculatrix was sister to a group that included series Spuriae and Tenuifoliae (Hall et al. Citation2000). I. speculatrix was also found to be outside of series Chinenses from our study. Further study is required to resolve the taxonomic placement of I. speculatrix between different series among the genus Iris.
Ethical statement
This research was conducted in accordance with the Legislation of Hong Kong Special Administrative Region. The sample collections did not cause any environmental problem.
Author contributions
Conceptualization: T.Y.S., D.T.W.L., and P.C.S.
Methodology: T.Y.S., K.H.W, B.L.H.K, H.Y.W., and G.W.C.B.
Data analysis: T.Y.S., K.H.W, and B.L.H.K.
Writing—original draft: T.Y.S.
Writing—review and editing: K.H.W, B.L.H.K, H.Y.W., G.W.C.B., D.T.W.L., and P.C.S.
Supervision: D.T.W.L. and P.C.S.
Acknowledgement
The authors would like to thank the Hong Kong Herbarium in collection and authentication of the local sample of Iris speculatrix Hance.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank (https://www.ncbi.nlm.nih.gov) with the accession number OK274247 (https://www.ncbi.nlm.nih.gov/nuccore/OK274247.1). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA796487, SRR17635354, and SAMN24866026, respectively.
Additional information
Funding
References
- Crespo MB, Martínez-Azorín M, Mavrodiev EV. 2015. Can a rainbow consist of a single colour? A new comprehensive generic arrangement of the ‘Iris sensu latissimo’clade (Iridaceae), congruent with morphology and molecular data. Phytotaxa. 232(1):864–78.
- Govaerts R, Nic Lughadha E, Black N, Turner R, Paton A. 2021. The World Checklist of Vascular Plants, a continuously updated resource for exploring global plant diversity. Sci Data. 8(1):215. [Cited as Iris speculatrix.]
- Guo J, Wilson CA. 2013. Molecular phylogeny of crested Iris based on five plastid markers (Iridaceae). Syst Bot. 38(4):987–995.
- Hall T, Tillie N, Chase W. 2000. A re-evaluation of the bulbous irises. Ann Bot. 58:123–126.
- Hance HF. 1875. Two additions to the Hong Kong Flora. J Bot. 13:196–197.
- Hu QM, T. l W, Xia NH, Xing FW, Lai P, Yip K-l. 2003. Rare and precious plants of Hong Kong. Hong Kong: Agriculture, Fisheries and Conservation Department, The Government of the Hong Kong Special Administrative Region
- Kang YJ, Kim S, Lee J, Won H, Nam GH, Kwak M. 2020. Identification of plastid genomic regions inferring species identity from de novo plastid genome assembly of 14 Korean-native Iris species (Iridaceae). PLoS One. 15(10):e0241178.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Mavrodiev EV, Martínez-Azorín M, Dranishnikov P, Crespo MB. 2014. At least 23 genera instead of one: The case of Iris L. sl (Iridaceae). PLoS One. 9(8):e106459.
- Tillie N, Chase MW, Hall T. 2000. Molecular studies in the genus Iris L.: a preliminary study. Ann Bot. 58:105–112.
- Wilson CA. 2020. Two new species in Iris series Chinenses (Iridaceae) from south-central China. PhytoKeys. 161:41–60.