Abstract
We present the complete mitochondrial genome of a Malaria vector Mosquito Anopheles sinensis Wiedemann, 1828 from South Korea. The mitochondrial genome is about 15,421 bp long and contains 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and an A-T rich region. The present data were compared with those from China with respect to PCG sequence differences, tRNA structure, gene order, and control region (CR) structure. An. sinensis mitochondrial genomes from northeast Asia share identical gene composition and gene order. In contrast, they have distinct differences in the CR within the range 8.75% (51/583 bp) to 9.95% (58/583 bp). The phylogenetic analysis showed that An. sinensis from South Korea was clustered together with those from China, but there existed distinct genetic distance between the two. Likewise, mitochondrial genome sequences from other Anopheles species were employed to infer phylogenetic relationships among the members of the genus Anopheles. This study further promotes the enrichment of An. sinensis mitochondrial genome data, providing useful information for their mitochondrial genetic differences along with geographical distances in northeast Asia.
Anopheles sinensis Wiedemann, 1828 is an oriental species widely distributed in South Korea, China, and Japan as well as in other North and Southeast Asian Countries (Lu Citation1997; Hwang Citation2007; Shin Citation2014). It belongs to the hyrcanus group (subgenus Anopheles) which is a complex species assemblage that includes more than 20 closely related species in South Korea and China (Lu Citation1997; Hwang et al. Citation2004, Citation2006; Ma and Xu Citation2005, Citation2015; Ree et al. Citation2005). It is a vector of Plasmodium vivax and certain worms such as Brugia malayi that causes lymphatic filariasis (Hasegawa Citation1913; Zhang et al. Citation1991; Ree and Hwang Citation2000; Zhou et al. Citation2012). Despite its disputable malaria vector capacity, An. sinensis is still incriminated as a competent vector for P. vivax malaria due to its abundant population size and wide distribution, which have led to occasional local malaria epidemics or outbreaks throughout history (Chai et al. Citation1994; Cho et al. Citation1994; Kho et al. Citation2001; Ree et al. Citation2001; Ren et al. Citation2015). Due to its location on a peninsula, An. sinensis populations in South Korea may not be affected by the gene flow from the continent or nearby islands, thus, making the populations suitable for studying genetic diversity (Jung et al. Citation2007). Likewise, analysis of the full mitochondrial genome from the Northeast Asian population would be appropriate to analyze differences in each sample collection (Hwang and Kim Citation1999). In this study, we first attempted to characterize the complete mitochondrial genome of An. sinensis collected from South Korea, which was then compared with Chinese ones.
An. sinensis samples were collected by using light traps in the high-risk areas of malaria, Mungi-ri, Tanhyoen-myeon, Paju-si, Kyonggi-do, South Korea (GPS. N37°49′54.72″, E126°43′24.7″). The specimen is kept in Kyungpook National University (KNU), Daegu, South Korea under the voucher no. LEGOA050001 (collector: Ui Wook Hwang, [email protected]) and total genomic DNA was isolated from tissues using a DNeasy Blood and Tissue Kit (Qiagen Co., Hilden, Germany) following the manufacturer’s protocol. The mitochondrial genome was amplified by the standard PCR method using the methods described by Luo et al. (Citation2016). The complete mitochondrial genome was amplified using 18 primer pairs designed by Zhang et al. (Citation2013). The amplified PCR products were checked on 1.0% agarose gel and purified using a QIAquick PCR Purification Kit (Qiagen Co., Hilden, Germany). Then, the amplicons were sequenced directly using an ABI Prism 3730 DNA sequencer (PerkinElmer, Waltham, MA) with a BigDye Termination Sequencing Kit. All alignments were performed in the Clustal X2 program (Larkin et al. Citation2007) and BioEdit 7.0.9 program (Hall Citation1999). The characterization of the protein-coding genes (PCGs), rRNAs, tRNAs, and control region (CR) was done using NCBI Basic Local Alignment Search Tool (BLAST) and tRNAscan-SE program (Chan and Lowe Citation2019). The PCG of the amino acid sequences used for the phylogenetic analysis was also aligned with Clustal X2 (Larkin et al. Citation2007). Phylogenetic relationships were inferred using the maximum-likelihood (ML) with IQ-TREE online webserver (Trifinopoulos et al. Citation2016). The phylogenetic tree was built with 1000 bootstrap replicates and substitution model mtART + F+I + G4.
The complete mitochondrial genome of An. sinensis (GenBank accession number: OK458560) is a double-stranded circular genome of 15,421 bp in size, containing 13 PCGs, 22 tRNA genes, two rRNA genes, and a CR. The whole nucleotide composition is 40.0% for A, 37.9% for T, 9.5% for G, and 12.7% for C, respectively, presenting an obvious A + T bias (77.9%). Most of genes of the mitochondrial genome are encoded on heavy strand except for four PCGs (ND1, ND4, ND4L, and ND5), nine tRNA genes (trnQ, trnC, trnY, trnS, trnF, trnH, trnP, trnL, and trnV), and two rRNA genes (rrnL and rrnS). Except for COI, ND4L, and ND1 starting with TCG, CTT, and CAT, respectively, other PCGs use ATG, ATA, or ATC as the initial codon. Seven PCGs (ND2, COI, APT8, ATP6, ND3, ND5, and ND4) stop with the complete termination codon TAA, and the rest have incomplete stop codon T. All tRNAs vary from 66 to 72 bp in length. The overall A + T content of 22 tRNA genes is 78.5%. Two rRNA genes were found: 16S rRNA with a length of 1268 bp and 12S rRNA with a length of 756 bp. The 16S rRNA is assumed to fill up the blanks between trnL and trnV, and the 12S rRNA is located between trnV and the CR. The CR is 583 bp long which has a higher A + T content (93.7%) than that of the whole mitochondrial genome (78.9%).
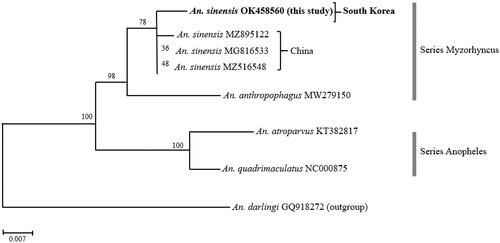
To determine the phylogenetic position of An. sinensis from South Korea within the Hyrcanus group, we reconstructed the ML tree based on the concatenated amino acid sequence alignment deduced from eight Anopheles mitochondrial genomes. We employed An. darlingi as an outgroup. As shown in , the major clades on the ML tree supported the monophylies of the two series Myzorhynchus (BP 98%) and Anopheles (BP 100%). An. sinensis from South Korea was clustered with Chinese ones, but there exhibited significant genetic distance between the two. Although An. anthropophagus appeared as a sister taxon of the monophyletic clade of An. sinensis, the two species are distinctly separated from each other.
Figure 1. A maximum-likelihood (ML) tree showing the mitochondrial genetic distance between South Korean and Chinese An. sinensis. The ML tree was reconstructed based on amino acid sequences of the 13 mitochondrial PCGs under the mtART + F+I + G4 model using the IQTREE program. A newly sequenced An. sinensis individual from South Korea was highlighted in bold.
Ethical approval
No specific permits were required for this study. The study did not involve endangered or protected species. Therefore, the ethics committee (Ministry of Environment, South Korea) deemed that approval was unnecessary.
Author contributions
Ashraf Akintayo Akintola conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared a figure, wrote the original draft, reviewed, and edited drafts of the manuscript. The experiments and data analysis were devised by Bia Park and Eun Hwa Choi, who then corrected critical drafts of the publication. Ui Wook Hwang conceived and designed the experiments and data analysis, reviewed and edited drafts of the manuscript approved the final draft.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete mitochondrial genome sequence of Anopheles sinensis from South Korea has been deposited in GenBank and openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/OK458560 under the accession no. OK458560. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA823808, SRR18650725, and SAMN27361716, respectively.
Additional information
Funding
References
- Chai IH, Lim GI, Yoon SN, Oh WI, Kim SJ, Chai JY. 1994. Occurrence of tertian malaria in a male patient who has never been abroad. Korean J Parasitol. 32(3):881–200.
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14.
- Cho SY, Kong Y, Park SM, Lee JS, Lim YA, Chae SL, Kho WG, Lee JS, Shim JC, Shin HK. 1994. Two vivax malaria cases detected in Korea. Korean J Parasitol. 32(4):281–284.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98.
- Hasegawa Y. 1913. Malaria in Korea. Chosen Igakkai Zasshi. 4:53–69.
- Hwang UW. 2007. Revisited ITS2 phylogeny of Anopheles (Anopheles) Hyrcanus group mosquitoes: reexamination of unidentified and misidentified ITS2 sequences. Parasitol Res. 101(4):885–894.
- Hwang UW, Kim W. 1999. General properties and phylogenetic utilities of nuclear ribosomal DNA and mitochondrial DNA commonly used in molecular systematics. Korean J Parasitol. 37(4):215–228.
- Hwang UW, Yong TS, Ree HI. 2004. Molecular evidence for synonymy of Anopheles yatsushiroensis and An. pullus. J Am Mosq Control Assoc. 20(2):99–104.
- Hwang UW, Tang LH, Kobayashi M, Yong TS, Ree HI. 2006. Molecular evidence supports that Anopheles anthropophagus from China and Anopheles lesteri from Japan are the same species. J Am Mosq Control Assoc. 22(2):324–326.2.0.CO;2]
- Jung J, Jung Y, Min GS, Kim W. 2007. Analysis of the population genetic structure of the malaria vector Anopheles sinensis in South Korea based on mitochondrial sequences. Am J Trop Med Hyg. 77(2):310–315.
- Kho WG, Chung JY, Hwang UW, Chun JI, Park YH, Chung WC. 2001. Analysis of polymorphic region of GAM-1 gene in Plasmodium vivax Korean isolates. Korean J Parasitol. 39(4):313–318.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23(21):2947–2948.
- Lu B. 1997. Fauna Sinica, Insecta. Vol. 9: Diptera, Culicidae II. Beijing: Science Press.
- Luo QC, Hao YJ, Meng FX, Li TJ, Ding YR, Hua YQ, Chen B. 2016. The mitochondrial genomes of Culex tritaeniorhynchus and Culex pipiens pallens (Diptera: Culicidae) and comparison analysis with two other Culex species. Parasit Vectors. 9(1):406.
- Ma Y, Xu J. 2005. The Hyrcanus group of Anopheles (Anopheles) in China (Diptera: Culicidae): species discrimination and phylogenetic relationships inferred by ribosomal DNA internal transcribed spacer 2 sequences. J Med Entomol. 42(4):610–619.2.0.CO;2]
- Ma Y, Xu J. 2015. Progress of taxonomic study on the Anopheline mosquitoes in China. Chin J Vector Biol Control. 26(5):433–438.
- Ree HI, Hwang UW. 2000. Comparative study on longevity of Anopheles sinensis in malarious and non-malarious areas in Korea. Korean J Parasitol. 38(4):263–266.
- Ree HI, Hwang UW, Lee IY, Kim TE. 2001. Daily survival and human blood index of Anopheles sinensis, the vector species of malaria in Korea. J Am Mosq Control Assoc. 17(1):67–72.
- Ree HI, Yong TS, Hwang UW. 2005. Identification of four species of the Anopheles hyrcanus complex (Diptera: Culicidae) found in Korea using species-specific primers for polymerase chain reaction assay. Med Entomol Zool. 56(3):201–205.
- Ren Z, Wang D, Hwang J, Bennett A, Sturrock HJW, Ma A, Huang J, Xia Z, Feng X, Wang J. 2015. Spatial–temporal variation and primary ecological drivers of Anopheles sinensis human biting rates in malaria epidemic-prone regions of China. PLOS One. 10(1):e0116932.
- Shin EH. 2014. Characteristics of malaria vector mosquitoes in the Republic of Korea. Public Health Week Rep KCDC. 7:813–818.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235.
- Zhang SQ, Zhang QJ, Cheng F, Wang LL, Pen GP. 1991. Threshold of transmission of Brugia malayi by Anopheles sinensis. J Trop Med Hyg. 94(4):245–250.
- Zhang NX, Zhang YJ, Yu G, Chen B. 2013. Structure characteristics of the mitochondrial genomes of Diptera and design and application of universal primers for their sequencing. Acta Entomol Sin. 56:398–407.
- Zhou SS, Zhang SS, Wang JJ, Zheng X, Huang F, Li WD, Xu X, Zhang H-W. 2012. Spatial correlation between malaria cases and water-bodies in Anopheles sinensis dominated areas of Huang-Huai plain, China. Parasit Vectors. 5:106.
