Abstract
Echeveria lilacina Kimnach & Moran Citation1980 is a succulent plant having ornamental and ecological importance. In this study, the first complete chloroplast genome of Echeveria lilacina, a species belonging to the Crassulaceae family, was characterized from the de novo assembly of Illumina NovaSeq 6000 paired-end sequencing data. The chloroplast genome of E. lilacina is 150,080 bp in length, which includes a large single-copy (LSC) region of 81,741 bp, a small single-copy (SSC) region of 16,747 bp, and a pair of identical inverted repeat regions (IRs) of 25,796 bp each. The genome annotation revealed a total of 138 genes, including 87 protein-coding genes, 41 transfer RNA (tRNA) genes, and 10 ribosomal RNA (rRNA) genes. The phylogenetic analysis with 15 complete chloroplast genome sequences including outgroup showed that E. lilacina formed the closest taxonomical relationship with Graptopetalum amethystinum in the Crassulaceae family.
Echeveria, a member of the Crassulaceae family, is a genus consisting of approximately 150 succulent species. Of them, Echeveria lilacina Kimnach & Moran Citation1980 (Kimnach and Moran Citation1980), also called Ghost Echeveria, has been often used as an ornamental plant and is known for an environmental host plant for butterflies (Ziegler and Escalante Citation1964). Echeveria lilacina is characterized by pale and silvery-gray fleshy leaves and pale pink or coral-pink flowers from late winter to early spring (López-Angulo et al. Citation2022). The habitat of E. lilacina ranges from semi-desert areas of west Texas to Argentina, across Mexico, Guatemala, and Central and South America (Palomino et al. Citation2021).
The sample of E. lilacina plant was collected from Nuevo León, Mexico (25°06′50.0″N, 100°11′50.0″W) in accordance with local regulation and permission of local authorities. The collected plant was deposited at National Agrobiodiversity Center at Rural Development Administration (http://genebank.rda.go.kr/distribGuide.do, Mun Sup Yoon, [email protected]), Korea under the genetic resource number IT317443. The specimen of E. lilacina under the voucher number SYCS101 was deposited at Natural Science Research Institute (https://www.syu.ac.kr/industry-academic-research/a-research-institute/natural-science-institute/#, contact person: Jae Hwan Lee, [email protected]), Sahmyook University, which is the affiliated organization of National Agrobiodiversity Center at Rural Development Administration, Korea and serves as the institute of agricultural genetic resources for succulent plants.
The leaves of E. lilacina were used to extract genomic DNA using a modified CTAB-based protocol, followed by DNA QC by Nanodrop and agarose gel electrophoresis. The genomic library for Illumina 150 bp paired-end (PE) was constructed according to the manufacturer’s recommendation using NEXTflex® Rapid DNA sequencing kit (Bioo Scientific, Austin, TX, USA). The chloroplast genome of E. lilacina was sequenced by NovaSeq 6000 (Illumina Inc., San Diego, CA, USA) and low-quality and adaptor sequences were removed by Trimmomatic (Bolger et al. Citation2014). The high-quality PE reads were assembled by CLC Genomics Workbench (ver. 10.0.1, CLC QIAGEN), followed by manual curation through PE reads mapping (Kim et al. Citation2015). Annotation of the complete chloroplast genome was performed with GeSeq and manual corrections (Tillich et al. Citation2017). The complete chloroplast genome sequence of E. lilacina was submitted to GenBank with the accession number MZ643065.
The complete chloroplast genome of E. lilacina was 150,080 bp in length with 37.88% of G + C content, comprising a large single copy (LSC) region of 81,741 bp, a small single copy (SSC) region of 16,747 bp, and a pair of inverted repeats (IRa and IRb) regions of 25,796 bp each. The genome contained 138 genes including 87 protein-coding genes, 41 tRNA genes, and 10 rRNA genes.
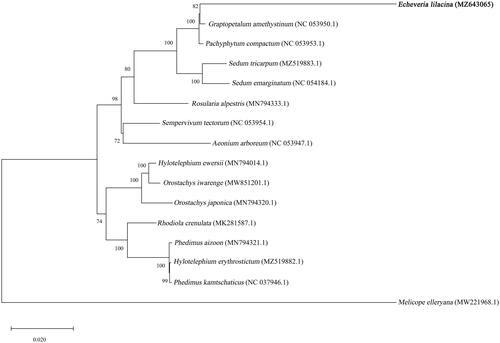
In order to investigate the evolutionary relationship, the complete chloroplast genome sequences of E. lilacina with 14 related species in Crassulaceae and one outgroup species, were aligned using ClustalW (ver. 2.1) (Larkin et al. Citation2007), followed by phylogenetic tree construction based on Maximum Likelihood (ML) algorithm with 1000 bootstraps using MEGA 10.2.5 (Kumar et al. Citation2018). The phylogenetic analysis showed the closest relationship of E. lilacina with Graptopetalum amethystinum in the family of Crassulaceae ().
Figure 1. Maximum-likelihood phylogenetic tree based on complete chloroplast genome sequences of E. lilacina (bold font) and 14 related species with one outgroup species, Melicope elleryana. The GenBank accession numbers are designated next to each species name. The values above branches are bootstrap percentages based on 1000 replicates.
Author contribution
SYN conceived the original structure of the project and reviewed the manuscript. JHL prepared the plant sample and SYS prepared the DNA of the sample. JRJ analyzed the data. GN interprets the data and prepared the manuscript. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
We appreciate the National Agrobiodiversity Center at Rural Development Administration, Korea for providing the plant of Echeveria lilacina Kimnach & Moran (Genetic resource No. IT317443).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the finding of this study are publically available in GenBank at http://www.ncbi.nlm.gov/genbank/, with reference number, MZ643065. The BioProject, BioSample, and SRA numbers are PRJNA751436, SAMN20518232, and SRR15321362, respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):889–2120.
- Kim K, Lee S-C, Lee J, Yu Y, Yang K, Choi B-S, Koh H-J, Waminal NE, Choi H-I, Kim N-H, et al. 2015. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655.
- Kimnach M, Moran R. 1980. Echeveria lilacina, a new species from Nuevo León, Mexico. Cact Succ J. 52(4):175–179.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23(21):2947–2948.
- López-Angulo G, Miranda-Soto V, López-Valenzuela JA, Montes-Avila J, Díaz-Camacho SP, Garzón-Tiznado JA, Delgado-Vargas F. 2022. Glucosidase inhibitory phenolics from Echeveria subrigida (B. L. Rob & Seaton) leaves. Nat Prod Res. 36(4):1058–1061.
- Palomino G, Martínez-Ramón J, Cepeda-Cornejo V, Ladd-Otero M, Romero P, Reyes-Santiago J. 2021. Chromosome number, ploidy level, and nuclear DNA content in 23 species of Echeveria (Crassulaceae). Genes. 12(12):1950.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Ziegler JB, Escalante T. 1964. Observations on the life history of Callophrys xami (Lycaenidae). J Lepid Soc. 18(2):85–89.
