Abstract
In this study, we collected plant material from Pseudotsuga sinensis in Guizhou, China, and sequenced it. The complete chloroplast genome consisted of 122,243 bp, including a large single-copy (LSC) region, a small single-copy (SSC) region, and two inverted repeat regions like those in P. sinensis var. wilsoniana. The GC content of P. sinensis and P. sinensis var. wilsoniana are 38.7% and 38.8%, respectively. The reconstructed phylogenetic tree reveals that P. sinensis was a sister species to P. sinensis var. wilsoniana. Hence, the availability of the chloroplast genome of P. sinensis will promote further phylogenetic studies of the family Pinaceae.
Pseudotsuga is a small genus of evergreen coniferous trees with four or five recognized species that are discontinuously distributed across East Asia and North America. It also is a rare relict plant unique to China and is classified as ‘endangered’ by the International Union for Conservation of Nature (IUCN; Li et al. 2021). Prior to this study, the NCBI database only contained chloroplast genomes for two Pseudotsuga species. The chloroplast genome size of most reported pinaceous species only ranges from 107 to 120 kb (Lin et al. Citation2010; Wu et al. Citation2011). To better address evolution-based questions regarding Pseudotsuga, we assembled and reported the first complete plastome of Pseudotsuga sinensis (Dode 1912).
Plant material from P. sinensis was collected from a village in Guizhou Province (26°35′11.04″N, 103°58′5.88″E). Voucher specimens (LPJ001) were deposited in the Herbarium of the Kunming Institute of Botany at the Chinese Academy of Sciences (KUN, 1519936, http://www.kun.ac.cn/). Total genomic DNA was collected from fresh leaves of an Aralia elata sample based on a previously suggested method (Li et al. Citation2013). Next, a shotgun library was constructed, and high-throughput sequencing was conducted on an Illumina Hiseq X-Ten sequencing platform. The clean data were assembled de novo using GetOrganelle v1.7.5.0 (Jin et al. Citation2020), and annotation was performed using GeSeq (Tillich et al. Citation2017). Subsequently, the annotations of P. sinensis were inspected manually using Geneious v9.0.2 (Kearse et al. Citation2012) and compared with P. sinensis var. wilsoniana. Finally, the annotated genomic sequence of P. sinensis was submitted to GenBank with the accession number MZ779058.
The complete chloroplast genome P. sinensis consists of 122,243 bp, including a large single-copy (LSC) region, a small single-copy (SSC) region, and two inverted repeating regions like those in P. sinensis var. wilsoniana. The GC content of P. sinensis is 38.7%, and it has a 22,332-bp inverted repeat region compared to the complete chloroplast genome of P. sinensis var. wilsoniana. The complete chloroplast genome of P. sinensis var. wilsoniana includes 111 genes, including 71 protein-coding sequences, 36 tRNA-encoding genes, and 4 rRNA-encoding genes. The complete chloroplast genome of P. sinensis var. wilsoniana includes 113 genes, including 73 protein-coding sequences, 36 tRNA-encoding genes, and 4 rRNA-encoding genes. The complete chloroplast genome of P. sinensis lacks the genes psbN and ycf12.
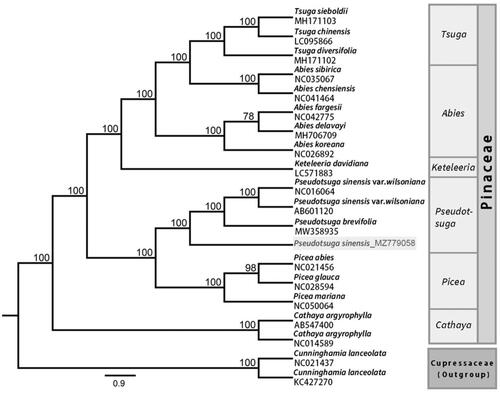
To obtain insight into the position of P. sinensis within the family Pinaceae, 19 other complete chloroplast genome sequences of Pinaceae were downloaded from the GenBank database, using two sequences of Cunninghamia lanceolata as outgroups (). Phylogenetic analyses were performed using maximum-likelihood (ML) in RAxML8.0 with 1000 bootstrap replicates (Stamatakis Citation2014). In the ML tree, most of the nodes had a 100% bootstrap value. The reconstructed phylogenetic tree revealed that P. sinensis is a sister species to Pseudotsuga brevifolia and P. sinensis var. wilsoniana. Although morphological data suggest that P. sinensis is more closely related to P. sinensis var. wilsoniana than to P. brevifolia, our analysis indicates close relationships between all three species. The ML tree based on complete chloroplast genome data did not provide further insights into the relationship between the three species. However, the addition of the whole chloroplast genome of P. sinensis to the NCBI database will aid in the phylogenetic reconstruction of the family Pinaceae.
Ethical approval
Fieldwork was approved and supervised by the Nature reserve service station for Pseudotsuga sinensis in Weining. Pseudotsuga sinensis is vulnerable (VU) according to IUCN Red list, so all the plant experiments were conducted according to policies research involving species at risk of extinction described in ‘the International Union for Conservation of Nature.’
Author contributions
PJ.L. and S. Z. designed the experiments and analyzed the data. WJ.L. wrote the original manuscript. J.L. collected the plant materials. T.F. and B.H. identified the materials. T.F., J.L., and S.Z. assisted with manuscript preparation. All authors have read and agreed to publish the manuscript.
Acknowledgment
The authors thank Q.L. from Yunnan Normal University for submitting the sequence.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://dataview.ncbi.nlm.nih.gov/] under accession no. MZ779058. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA755471, SRR15497848, and SAMN20824202 respectively.
Additional information
Funding
References
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li J, Wang S, Jing Y, Wang L, Zhou S. 2013. A modified CTAB protocol for plant DNA extraction. Chinese Bull Bot. 48(1):72–78.
- Li LL, Xu YZ, Zhou TY, Yang ZZ, Huang HD, Gu ZR, Jiang MX. 2021. Population structure and community characteristics of Pseudotsuga sinensis Dode in Badagongshan, Hunan Province, China. Plant Sci J. 39(2):111–120.
- Lin CP, Huang JP, Wu CS, Hsu CY, Chaw SM. 2010. Comparative chloroplast genomics reveals the evolution of Pinaceae genera and subfamilies. Genome Biol Evol. 2:504–517.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wu CS, Lin CP, Hsu CY, Wang RJ, Chaw SM. 2011. Comparative chloroplast genomes of Pinaceae: insights into the mechanism of diversified genomic organizations. Genome Biol Evol. 3:309–319.