Abstract
Habenaria dentata is a rare species with high ornamental value in China. In this study, we report the complete chloroplast (cp) genome of H. dentata using the Illumina sequencing data. The total genome of H. dentata is 153,682 bp in length and the GC content is 36.62%, with a pair of inverted repeats (IRs) regions of 26,339 bp each, a large single-copy (LSC) region of 83,963 bp and a small single-copy (SSC) region of 17,041 bp. The cp genome encoded 133 genes, including 87 protein-coding genes (PCG), eight rRNA genes, and 38 tRNA genes. The maximum-likelihood phylogenetic analysis based on 12 cp genomes showed that H. dentata was sister to Habenaria chejuensis and Habenaria ciliolaris. This work will be valuable for genetic and phylogenetic studies on H. dentata.
Habenaria is one of the largest genera in the Orchidaceae family, including about 876 species and mainly distributed in tropical and subtropical areas (Govaerts et al. Citation2013). According to Flora of China, 58 species of Habenaria were reported in China (Zhang et al. Citation2016). Habenaria dentata (Sw.) Schltr 1919 is a species belonging to the Habenaria genera. It grows on the forest understory or gully sides of mountain slopes at an altitude of 190–2300 m (Chen et al. Citation2009). H. dentata has high ornamental value, and its tubers can be used medicinally, with the effect of diuretic, swelling, waist strength and kidney, treating lumbago, hernia and other diseases. However, due to its slow growth, special ecological requirements, difficulty in reproduction and long-term exploitation, the species is now very rare. Habenaria dentata is classified as a second-level key protected plant in the ‘National List of Key Protected Wild Plants (Second Batch)’ (http://www.iplant.cn/rep/prot/Habenaria%20dentata). To better protect H. dentata and provide significant genomic resources in the further study of Orchidaceae, we reported its chloroplast (cp) genome first and analyzed its structural characteristics and phylogenetic relationship.
Fresh leaves of H. dentata were collected at the Yachang Orchid National Nature Reserve, Baise, Guangxi, China (24°50′55″N, 106°21′5″E), and the specimens were preserved in the herbarium of the Guangxi Institute of Botany (contact: Shengfeng Chai, e-mail: [email protected], voucher number: SFC-20201015012). The sample collection was permitted by the Yachang Orchid National Nature Reserve Management Center, and the sampling process was followed by the Regulations of the People's Republic of China on the Protection of Wild Plants. Total genomic DNA was extracted using a modified CTAB method (Doyle And Doyle Citation1987), followed by double-end sequencing using the Illumina HiSeq 2000 platform, and the cp genome was assembled from scratch using the GetOrganelle script (Jin et al. Citation2018). Finally, we obtained the complete cp genome of H. dentata and used the online annotation software Geseq (Tillich et al. Citation2017) and CpGAVAS (Liu et al. Citation2012), using H. pantlingiana (NC_026775) as the reference genome, for cp genome annotation was spliced. Circular genome maps were drawn using the online tool OGDRAW (Lohse et al. Citation2007), and after proofreading and correction, the whole cp genome was submitted to GenBank (NCBI accession number: OK012095).
The complete cp genome sequence of H. dentata was 153,682 bp in length and showed a typical quadripartite structure, consisting of the large single-copy (83,963 bp) and small single-copy (17,041 bp) separated by a pair of inverted repeat (IR) (26,339 bp). It contains 133 protein-coding genes, 38 tRNA genes, and eight rRNA genes. The overall GC content of H. dentata cp genome was 36.62%, and in the LSC, SSC, and IR regions were 34.15, 28.96, and 43.04%, respectively. By comparing the cp genomes of H. dentata, we found that H. dentata is very similar to the cp genomes of previously reported Habenaria species in terms of gene content, gene order, and GC content.
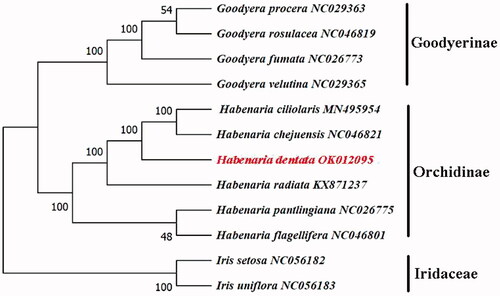
We performed a phylogenetic analysis with 12 complete cp genomes of Orchidaceae and two species of Iridaceae as outgroups. The GenBank accession numbers of the sequences used in this paper are shown in . Sequence alignment was performed using MAFFT (Katoh and Standley Citation2013), and a maximum-likelihood (ML) tree in the GTRGAMMA model was constructed using RAxML 8.2.12 (Stamatakis Citation2014), and the phylogenetic tree showed that H. dentata was clustered with Habenaria chejuensis and Habenaria ciliolaris with 100% bootstrap support (). In conclusion, the complete cp genome of H. dentata was decoded for the first time, which will facilitate the species identification, molecular biology, and phylogenetic studies of H. dentata. This cp genome will provide important clues to better understand the phylogeny and biodiversity of Habenaria.
Author contributions
S.C. conceived the research; S.C. collected samples; J.Z., Y.Y., J.T., Y.L., and H.L. analyzed and interpreted data; J.Z. wrote the manuscript; S.C., Y.Y., and J.Z. revised the manuscript. All authors approved the final version of the article and agreed to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The specimens of Habenaria dentata were preserved in the herbarium of the Guangxi Institute of Botany (voucher number: SFC-20201015012). The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OK012095. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA759395, SRR15685573, and SAMN21166999, respectively. Tree file of 12 species and genes for phylogenetic analysis were deposited at Figshare: https://doi.org/10.6084/m9.figshare.16910296.
Additional information
Funding
References
- Chen SC, Liu ZJ, Zhu GH, Lang KY, Tsi ZH, Luo YB, Jin XH, Cribb PJ, Wood JJ, Gale SW, et al. 2009. Flora of China. Vol. 25. Beijing, China: Science Press.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:1–15.
- Govaerts R, Bemet P, Kratochvil K, Gerlach G, Carr G, Alrich P, Pridgeon AM, Pfahl J, Campacci MA, Baptista DH, et al. 2013. World checklist of Orchidaceae. London (UK): Botanic Gardens, Kew.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv.256479.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715–717.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52(5–6):267–274.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Zhang WL, Gao JY, Liu Q. 2016. Habenaria chlorina, a newly recorded species of Orchidaceae form China. Plant Sci J. 34(1):18–20.