Abstract
Syzygium samarangense (Blume) Merr. et Perry, 1938, commonly known as wax apple, is a Myrtaceae species that is known for its unique fruit shape, flavorful and colorful fruits, medicinal value and increasing economic relevance. In this study, we reported the complete chloroplast genome sequence of S. samarangense. The complete genome is 159,109 bp in length with a quadripartite structure containing two single copy regions, a Large Single Copy region (LSC, 88,155 bp) and a Small Single Copy region (SSC, 18,796 bp) separated by Inverted Repeat regions (IRs, 26,079 bp). The GC content was 37.0%. It encoded 126 genes, including 81 protein-coding genes, 37 transfer RNA genes, and 8 ribosomal RNA genes. The phylogenetic relationships of 20 species inferred that all Syzygium species formed a single cluster belonging to Syzygieae tribe. Our results offer insights into the evolutionary relationship of S. samarangense within Myrtaceae, indicating a closer relationship between S. samarangense and S. forrestii.
Wax apple (Syzygium samarangense (Blume) Merr. et Perry, 1938) is a Myrtaceae species native to Malaya to the Andaman and the Nicobar Islands (Morton Citation1987), and has become economically important in Asia such as Malaysia, Indonesia, Thailand, and China (Vara-ubol et al. Citation2006; Moneruzzaman et al. Citation2015). The extract of S. samarangense fruits has therapeutic value against diabetic progression (Shen et al. Citation2013; Shen and Chang Citation2013) and human colon cancer (Simirgiotis et al. Citation2008). By nuclear ribosomal DNA and chloroplast markers, the evolutionary history of some groups of Myrtaceae such as Myrteae (Vasconcelos et al. Citation2017; Bernardes et al. Citation2018), Eugenia (Mazine et al. Citation2014) and Myrcia (Staggemeier et al. Citation2015) has been investigated. However, there are only few phylogenetic and evolutionary studies based on chloroplast genomes in the case of Syzygieae (Asif et al. Citation2013; Zhang et al. Citation2019). In this study, the complete chloroplast genome of S. samarangense was assembled and subjected to phylogenetic analysis, to provide more information on better understanding about Myrtaceae family.
The specimen of S. samarangense was deposited at the Field GenBank for wax apple of Fujian Academy of Agricultural Sciences, Fujian province, China (http://www.faas.cn/cms/html/gsyjs/index.html; Coordinates: 26°7′53″N; 119°20′6″E; Jiahui Xu, [email protected]) under the voucher number GPLWFJGSS0058. Total genomic DNA was extracted from the fresh leaves following the manufacturer’s protocol of Hi-DNAsecure Plant Kit (Tiangen Biotech, Beijing, China). The qualified libraries were constructed and performed with the Illumina Hiseq 2500 platform at Genepioneer Biotechnologies Inc. (Nanjing, China), and all the raw reads were trimmed by Fastqc. The complete chloroplast genome was assembled by NOVOPlasty (Dierckxsens et al. Citation2017) until a circular genome formed, with published S. cumini species (NCBI Accession No. GQ870669.3) as references. The chloroplast genome annotation was performed using the CpGAVAS (Liu et al. Citation2012) and then manually corrected. The complete chloroplast genome of S. samarangense was submitted to NCBI (Accession No. MW698694).
The complete chloroplast genome of S. samarangense was 159,109 bp in length, with a typical quadripartite circular structure containing a pair of inverted repeat regions (IR; 26,079 bp), large single copy region (LSC; 88,155 bp) and small single copy region (SSC; 18,796 bp). The GC content of the S. samarangense chloroplast genome was 37.0%. A total of 126 genes were predicted, including 81 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. There were 17 intron-containing genes identified, in which eight are distributed over LSC region, one in SSC region, and eight in IR regions. Besides, 34 long repeats sequences were detected. Among them, 18 repeats were found in the LSC (11), SSC (4), IR (3) regions, and the remaining 16 repeats were all across two regions.
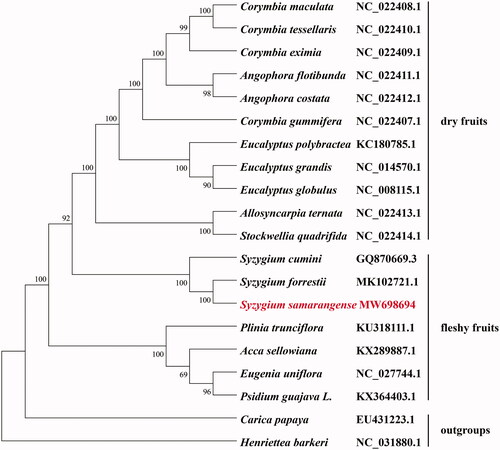
To investigate the evolutionary relationship of S. samarangense within Myrtaceae, eighteen representative species of Myrtaceae family were selected, with two species (Carica papaya and Henriettea barkeri) as outgroups. Complete chloroplast genome sequences were initially aligned using MAFFT (Katoh and Standley Citation2013) and then visualized and manually adjusted using BioEdit (Hall Citation1999). The maximum-likelihood analyze was conducted by using RaxML-HPC2 on TG ver. 7.2.8 on the Cipres web server (Miller et al. Citation2010) to reconstruct the phylogenetic tree. The results showed that eleven species with dry fruits were clustered together to Eucalypteae tribe (Thornhill et al. Citation2015). Seven species with fleshy fruits were clustered together, in which S. samarangense, S. forrestiiand and S. cumini formed a single cluster belonging to Syzygieae tribe (). This result supported the position that the emergence of fleshy fruit in the Syzygieae tribe occurred from a dry ancestral fruit form (Balbinott et al. Citation2022). Our results will provide valuable data for further phylogenomic study of Myrtaceae species.
Ethical approval
In this study, the plant materials (Syzygium samarangense ‘Tub Ting Jiang’) were collected and studied in accordance with the Agriculture Industry Standard of China ‘Descriptors standard for tropical crops germplasm-Wax apple (NY/T 3810-2020)’ and relevant Chinese regulations.
Author contributions
XW and JX obtained the funding support. LZ and JX conceived and designed the experiments. XW, LX and XZ collected and prepared plant materials. XW, LX and LL performed the experiments. XW and LL analyzed and interpreted the data. XW drafted the manuscript. LL revisited it critically. JX and LZ final approve the manuscript to be published. All authors agree to be accountable for all aspects of this work.
Disclosure statement
No potential conflict interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession MW698694. Raw Illumina data were deposited in the NCBI Sequence Read Archive (SRA: SRR17364499, BioProject: PRJNA607337, and Bio-Sample: SAMN14130528).
Additional information
Funding
References
- Asif H, Khan A, Iqbal A, Khan IA, Heinze B, Azim MK. 2013. The chloroplast genome sequence of Syzygium cumini (L.) and its relationship with other angiosperms. Tree Genet Genom. 9(3):977–877.
- Balbinott N, Rodrigues NF, Guzman FL, Turchetto-Zolet AC, Margis R. 2022. Perspectives in Myrtaceae evolution from plastomes and nuclear phylogenies. Genet Mol Biol. 45(1):e20210191.
- Bernardes C, Tuler AC, Ferreira A, Carvalho MS, Nogueira AM, Ferreira M. 2018. Transferability of Psidium microsatellite loci in Myrteae (Myrtaceae): a phylogenetic signal. Euphytica. 214(7):105.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Hall T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Sympos Series. 41:95–98.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Mazine FF, Souza VC, Sobral M, Forest F, Lucas E. 2014. A preliminary phylogenetic analysis of Eugenia (Myrtaceae: Myrteae), with a focus on neotropical species. Kew Bull. 69(2):9497.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. 2010 Gateway Computing Environments Workshop (GCE). New Orleans: IEEE. p. 1–8.
- Moneruzzaman KM, Sharif HA, Normaniza O, Nashriyah M, Nasrulhaq BA. 2015. Growth, yield and postharvest quality of wax apple as affected by naphthalene acetic acid application. Revista Brasileira De Fruticultura. 37:410–422.
- Morton JF. 1987. Java Apple. In: Morton JF, editor. Fruits of warm climates. Winterville: Creative Resource Systems, Inc.; p. 381–382.
- Shen SC, Chang WC. 2013. Hypotriglyceridemic and hypoglycemic effects of Vescalagin from Pink wax apple [Syzygium samarangense (Blume) Merrill and Perry cv. Pink] in high-fructose diet-induced diabetic rats. Food Chem. 136(2):858–863.
- Shen SC, Chang WC, Chang CL. 2013. An extract from wax apple (Syzygium samarangense (Blume) Merrill and Perry) effects glycogenesis and glycolysis pathways in tumor necrosis factor-α-treated FL83B mouse hepatocytes. Nutrients. 5(2):455–467.
- Simirgiotis MJ, Adachi S, To S, Yang H, Reynertson KA, Basile MJ, Gil RR, Weinstein IB, Kennelly EJ. 2008. Cytotoxic chalcones and antioxidants from the fruits of a Syzygium samarangense (Wax Jambu). Food Chem. 107(2):813–819.
- Staggemeier VG, Diniz-Filho JAF, Forest F, Lucas E. 2015. Phylogenetic analysis in Myrcia section Aulomyrcia and inferences on plant diversity in the Atlantic rainforest. Ann Bot. 115(5):747–761.
- Thornhill AH, Ho SYW, Külheim C, Crisp MD. 2015. Interpreting the modern distribution of Myrtaceae using a dated molecular phylogeny. Mol Phylogenet Evol. 93:29–43.
- Vara-ubol S, Chambers E, IV, Kongpensook V, Oupadissakoon C, Yenket R, Retiveau A. 2006. Determination of the sensory characteristics of rose apples cultivated in Thailand. J Food Science. 71(7):S547–S552.
- Vasconcelos TNC, Proença CEB, Ahmad B, Aguilar DS, Aguilar R, Amorim BS, Campbell K, Costa IR, De-Carvalho PS, Faria JEQ, et al. 2017. Myrteae phylogeny, calibration, biogeography and diversification patterns: increased understanding in the most species rich tribe of Myrtaceae. Mol Phylogenet Evol. 109:113–137.
- Zhang X-F, Wang J-H, Wang H-X, Zhao K-K, Zhu Z-X, Wang H-F. 2019. Complete plastome sequence of Syzygium forrestii Merr. et Perry (Myrtaceae): an endemic species in China. Mitochondr DNA Part B. 4(1):126–127.