Abstract
The complete mitochondrial (mt) genome of Leptomantella tonkinae (Hebard, 1920) was 15,527 bp in length and contained 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs, and one control region. The gene arrangement of mt genome of L. tonkinae was identical to the primitive mantis. The overall AT content of the mt genome was 74%. In ML and BI phylogenetic analyses, the monophyly of Leptomantellidae was robustly supported and the clade of Leptomantellidae is a sister clade to the group of (Gonypetidae+(Leptomantellidae+(Amorphoscelidae+Nanomantidae))).
Praying mantises are a major group of predatory insects, with over 2500 species/subspecies in 29 families and 60 subfamilies according to the website http://Mantodea.SpeciesFile.org (Otte et al. 2021). Leptomantellidae is originally included in Iridopterygidae as a genus (Leptomantella), which has recently been promoted to a new family that included four genera (Ehrmann Citation2002; Wieland 2013; Schwarz and Roy Citation2019). Leptomantellids can be distinguished from other mantises because of some special characteristics including juxta-ocular bulges distinct, rounded; wings mostly hyaline or subhyaline; and the presence of four discoidal spines (Schwarz and Roy Citation2019). However, only one complete mitochondrial (mt) genome of Leptomantellidae (Leptomantella albella GenBank: KJ463364) was available in NCBI (Wang, Yu, et al. Citation2016a), which hindered the phylogenetic relationship of Leptomantellidae. Hence, to enrich the molecular dataset of the Leptomantellidae, we sequenced the complete mt genome of Leptomantella tonkinae (Hebard, 1920) (Mantodea: Leptomantellidae). The mt genome was deposited in GenBank with accession no. OK480879. The BI and ML phylogenetic trees were constructed by using the 13 PCGs of mt genomes, which aimed to discuss the phylogenetic relationship of Leptomantellidae.
Leptomantella tonkinae is not a protected insect species, so no permission for scientific research in China is needed. Our sample (GXJX20170714-1) was collected from Jinxiu Yao Autonomous County, Guangxi Zhuang Autonomous Region, China (24.13°N, 110.18°E). Based on the taxonomic system of Otte, the sample was identified as L. tonkinae by its morphometric features and was stored at −40 °C in the Animal Specimen Museum, College of Chemistry and Life Sciences, Zhejiang Normal University, China. Whole genomic DNA was extracted from leg muscle tissue using an Ezup Column Animal Genomic DNA Purification Kit (Sangon Biotech Company, Shanghai, China) and stored at Zhang’s lab (http://mypage.zjnu.edu.cn/ZJY3/zh_CN/index.htm, Zhang JY, [email protected]). The mt genome was amplified with several universal primers as described in Zhang et al. (Citation2018a). To fill the gaps in previously obtained sequences, some species-specific primers were designed using Primer Premier 5.0 (Lalitha Citation2000). Finally, the DNA fragments were checked and assembled using DNASTAR Package v.7.1 (Burland Citation2000).
The complete mt genome of L. tonkinae was 15,527 bp in length and included 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs), and one control region (681 bp). The nucleotide composition of L. tonkinae was shown to be A: 37.9%, C: 15.7%, G: 10.3%, and T: 36.1%, with an overall AT% content of 74%. Furthermore, the AT-skew and GC-skew of the mt genome were 0.025 and −0.207, respectively, which was similar to the published mt genome of L. albella (Wang, Yu, et al. Citation2016a). Additionally, a 32 bp non-coding region located between trnP and ND6 was also found in L. albella (Wang, Yu, et al. Citation2016a). Based on the mt genome, the genetic distance with the Kimura 2-pweremeters model between L. albella and L. tonkinae was calculated as 0.056 by MEGA7.0 (Kumar et al. Citation2016). This gene arrangement was identical to the primitive insect mt genomes (e.g. Ayivi et al. Citation2021; Guan et al. Citation2021; Xu et al. Citation2021; Zhang et al. Citation2021).
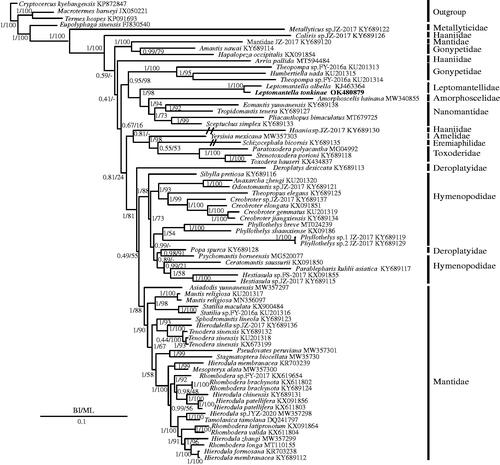
To discuss the phylogenetic position of L. tonkinae, one newly sequenced mantis mt genome as well as 68 previously sequenced mantis mt genomes were used in phylogenetic analyses (Cameron et al. Citation2006; Song et al. Citation2016; Wang, Yu, et al. Citation2016a; Wang, Hou, et al. 2016b; Ye et al. Citation2016; Tian et al. Citation2017; Zhang and Ye Citation2017; Zhang et al. Citation2018a, Citation2018b, Citation2018c, Citation2018d; Jia et al. Citation2019; Shi et al. Citation2019; Yan and Lin Citation2019; Zhang et al. 2019; Guan et al. Citation2020; Wang et al. Citation2020; Xu et al. Citation2021). In addition, two cockroach species, Eupolyphaga sinensis and Cryptocercus kyebangensis (Zhang et al. 2010) and two termites, Termes hospes (Dietrich and Brune 2016) and Macrotermes barneyi (Wei et al. Citation2012) were chosen as the outgroups. Based on the 13 PCGs, two types of phylogenetic trees were constructed using maximum-likelihood (ML) (Stamatakis Citation2014) and Bayesian inference (BI) (Huelsenbeck et al. Citation2001) methods via the MrBayes with a GTR + I+G model and RAxML 8.2.0 with a GTRGAMMAI model, respectively. In BI and ML trees (), Leptomantella tonkinae was a sister clade to L. albella, and Leptomantellidae was a sister clade to the clade of (Amorphoscelidae + Nanomantidae). According to the results of Schwarz and Roy (Citation2019), they supported the promotion of Leptomantella to the family level. This was consistent with previous studies that supported the monophyly of the families Nanomantidae, Toxoderidae, and Mantidae (Zhang et al. Citation2018a, Citation2018b, Citation2018c, Citation2018d, 2019; Xu et al. Citation2021). However, Hymenopodidae and Deroplatyidae were shown to be paraphyletic groups and the monophyly of the families Haaniidae and Deroplatyidae was not supported, which was also found in the results of Zhang et al. (Citation2018a, Citation2018d). Hence, to explore these discrepancies, more mt genomes should be sequenced in the future.
Figure 1. Phylogenetic tree of the relationships among 69 species of Mantodea inferred from BI analysis (A) and ML analysis (B) with two cockroaches (Eupolyphaga sinensis, Cryptocercus kyebangensis), and two termite (Termes hospes, Macrotermes barneyi) species chosen as outgroups. The GenBank accession numbers of all species are shown in the figure.
Author contributions
All authors were involved in the conception and design, or analysis and interpretation of the data; Lin YJ, Zhao YY, Yang YM, Jin WT, and Cai LN were involved in the drafting of the paper; Storey KB, Zhang JY, and Yu DN were involved in revising it critically for intellectual content; and all authors were involved in the final approval of the version to be published; and that all authors agree to be accountable for all aspects of the work.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the article.
Data availability statement
The mitochondrial genome data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/OK480879 under the accession no. OK480879. The mt genome was obtained by the Sanger method, so no associated ‘BioProject’, ‘SRA’, and ‘Bio-Sample’ numbers should be shown.
Additional information
Funding
References
- Ayivi SPG, Tong Y, Storey KB, Yu DN, Zhang JY. 2021. The mitochondrial genomes of 18 new Pleurosticti (Coleoptera: Scarabaeidae) exhibit a novel trnQ-NCR-trnI-trnM gene rearrangement and clarify phylogenetic relationships of subfamilies within Scarabaeidae. Insects. 12(11):1011.
- Burland TG. 2000. DNASTAR’s Lasergene sequence analysis software. Methods Mol Biol. 132:71–91.
- Cameron SL, Barker SC, Whiting MF. 2006. Mitochondrial genomics and the new insect order Mantophasmatodea. Mol Phylogenet Evol. 38(1):274–279.
- Dietrich C, Brune A. 2016. The complete mitogenomes of six higher termite species reconstructed from metagenomic datasets (Cornitermes sp., Cubitermes ugandensis, Microcerotermes parvus, Nasutitermes corniger, Neocapritermes taracua, and Termes hospes). Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):3903–3904.
- Ehrmann R. 2002. Mantodea: Gottesbeterinnen der welt. Germany: Natur und Tier-Verlag.
- Guan JY, Jia YY, Zhang ZY, Cao SS, Ma JL, Zhang JY, Yu DN. 2020. The complete mitochondrial genome of Xanthomantis bimaculata (Mantodea: Iridopterygidae) and its phylogeny. Mitochondrial DNA B. 5(3):3097–3099.
- Guan JY, Shen SQ, Zhang ZY, Xu XD, Storey KB, Yu DN, Zhang JY. 2021. Comparative mitogenomes of two Coreamachilis species (Microcoryphia: Machilidae) along with phylogenetic analyses of Microcoryphia. Insects. 12(9):795.
- Huelsenbeck J, Ronquist PJ, Fredrik F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Jia YY, Zhang LP, Xu XD, Dai XY, Yu DN, Storey KB, Zhang JY. 2019. The complete mitochondrial genome of Mantis religiosa (Mantodea: Mantidae) from Canada and its phylogeny. Mitochondrial DNA B Resour. 4(2):3797–3799.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lalitha S. 2000. Primer Premier 5. Biotech Softw Internet Rep. 1(6):270–272.
- Otte D, Spearman L, Stiewe MBD. 2021. Mantodea species file online. Version 5.0/5.0; [accessed 2021 Oct 20]. http://Mantodea.SpeciesFile.org.
- Schwarz CJ, Roy R. 2019. The systematics of Mantodea revisited: an updated classification incorporating multiple data sources (Insecta: Dictyoptera). Ann Soc Entomol Fr. 55(2):101–196.
- Shi Y, Liu QP, Luo L, Yuan ZL. 2019. Characterization of the complete mitochondrial genome sequence of Asiadodis yunnanensis (Mantidae: Choeradodinae) and phylogenetic analysis. Mitochondrial DNA B Resour. 4(2):2826–2827.
- Song N, Li H, Song F, Cai W. 2016. Molecular phylogeny of Polyneoptera (Insecta) inferred from expanded mitogenomic data. Sci Rep. 6(1):1–10.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tian X, Liu J, Cui Y, Dong P, Zhu Y. 2017. Mitochondrial genome of one kind of giant Asian mantis, Hierodula formosana (Mantodea: Mantidae). Mitochondrial DNA A DNA Mapp Seq Anal. 28(1):11–12.
- Wang Y, Cheng Y, Zhang Y. 2020. Characterization of the complete mitochondrial genome of the praying mantis Rhombodera longa (Mantodea: Mantidae) including a phylogenetic analysis. Mitochondrial DNA B. 5(2):1582–1583.
- Wang S, Hou F, Cao J, Peng C, Guo J. 2016. The complete mitochondrial genome of the Statilia maculata (Mantodea: Mantidae). Mitochondrial DNA B Resour. 1(1):860–861.
- Wang TT, Yu PP, Ma Y, Cheng HY, Zhang JY. 2016. The complete mitochondrial genome of Leptomantella. albella (Mantodea: Iridopterygidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):465–466.
- Wei SJ, Ni JF, Yu ML, Shi BC. 2012. The complete mitochondrial genome of Macrotermes barneyi Light (Isoptera: Termitidae). Mitochondrial DNA A. 23(6):426–428.
- Wieland F. 2013. The phylogenetic system of Mantodea (Insecta: Dictyoptera). Göttingen: Universitätsverlag Göttingen.
- Xu KK, Chen QP, Ayivi SPG, Guan JY, Storey KB, Yu DN, Zhang JY. 2021. Three complete mitochondrial genomes of Orestes guangxiensis, Peruphasma schultei, and Phryganistria guangxiensis (Insecta: Phasmatodea) and their phylogeny. Insects. 12(9):779.
- Xu XD, Guan JY, Zhang ZY, Cao YR, Storey KB, Yu DN, Zhang JY. 2021. Novel tRNA gene rearrangements in the mitochondrial genomes of praying mantises (Mantodea: Mantidae): translocation, duplication and pseudogenization. Int J Biol Macromol. 185:403–411.
- Yan S, Lin YZ. 2019. Characterization of the complete mitochondrial genome of the praying mantis Mekongomantis quinquespinosa (Mantodea, Mantidae). Mitochondrial DNA B Resour. 4(2):3280–3281.
- Ye F, Lan X, Zhu W, You P. 2016. Mitochondrial genomes of praying mantises (Dictyoptera, Mantodea): rearrangement, duplication, and reassignment of tRNA genes. Sci Rep. 6(1):25634.
- Zhang LP, Cai YY, Yu DN, Storey KB, Zhang JY. 2018a. Gene characteristics of the complete mitochondrial genomes of Paratoxodera polyacantha and Toxodera hauseri (Mantodea: Toxoderidae). Peerj. 6:e4595.
- Zhang LP, Cai YY, Yu DN, Storey KB, Zhang JY. 2018b. The complete mitochondrial genome of Psychomantis borneensis (Mantodea: Hymenopodidae). Mitochondrial DNA B. 3(1):42–43.
- Zhang ZY, Guan JY, Cao YR, Dai XY, Storey KB, Yu DN, Zhang JY. 2021. Mitogenome analysis of four Lamiinae species (Coleoptera: Cerambycidae) and gene expression responses by Monochamus alternatus when infected with the parasitic nematode, Bursaphelenchus mucronatus. Insects. 12(5):453.
- Zhang LP, Ma Y, Yu DN, Storey KB, Zhang JY. 2019. The mitochondrial genomes of Statilia maculata and S. nemoralis (Mantidae: Mantinae) with different duplications of trnR genes. Int J Biol Macromol. 121:839–845.
- Zhang YY, Xuan WJ, Zhao JL, Zhu CD, Jiang GF. 2010. The complete mitochondrial genome of the cockroach Eupolyphaga sinensis (Blattaria: Polyphagidae) and the phylogenetic relationships within the Dictyoptera. Mol Biol Rep. 37(7):3509–3516.
- Zhang HL, Ye F. 2017. Comparative mitogenomic analyses of praying Mantises (Dictyoptera, Mantodea): origin and evolution of unusual intergenic gaps. Int J Biol Sci. 13(3):367–382.
- Zhang L-P, Yu D-N, Storey KB, Cheng H-Y, Zhang J-Y. 2018c. Data for praying mantis mitochondrial genomes and phylogenetic constructions within Mantodea. Data Brief. 21:1277–1285.
- Zhang LP, Yu DN, Storey KB, Cheng HY, Zhang JY. 2018d. Higher tRNA gene duplication in mitogenomes of praying mantises (Dictyoptera, Mantodea) and the phylogeny within Mantodea. Int J Biol Macromol. 111:787–795.
