Abstract
Viola selkirkii, belonging to the genus Viola, has heart-shaped leaves and pale purple flowers, and it is widely distributed in the Northern Hemisphere, including Europe, North America, and Asia. In this study, the plastid genome of V. selkirkii was sequenced and phylogenetic analysis was performed on 11 Viola plastid genomes. The length of the plastid genome length of V. selkirkii was 156,774 bp, and it was identified as having a typical quadripartite structure with a large single-copy region (85,930 bp), a small single-copy region (17,982 bp), and two inverted repeat regions (26,431 bp each). A phylogenetic analysis was conducted with 77 protein-coding genes from the complete plastid genomes of 11 Viola and nine Salicaceae species; the complete plastid genome of Erythroxylum novogranatense was used as an outgroup. Viola formed a monophyletic clade, and V. selkirkii was closely related to V. ulleungdoensis. These results contribute to the clear identification of the phylogenetic position of V. selkirkii in Viola.
Violaceae Batch., which contains Viola selkirkii Pursh ex Goldie 1822, includes herbaceous, shrubs, and small shrubs and consists of 22 genera, and approximately 1000–1100 species (Wahlert et al. Citation2014). The largest genus, Viola L., contains approximately 580–620 species, and most of them are distributed in the Northern Hemisphere (Clausen Citation1964; Yoo and Jang Citation2010; Wahlert et al. Citation2014; Cheon et al. Citation2020). V. selkirkii is widely distributed in the Northern Hemisphere, including Europe, North America, and Asia, and its habitat is shaded, cold mountain forests and moist soils at high latitudes (Brainerd Citation1921; Russell Citation1956; Kim Citation1986). V. selkirkii is recognized as a member of Viola sect. Plagiostigma ser. Estolonosae (Marcussen et al. Citation2012). A morphological study of V. selkirkii was conducted by Russell (Citation1956). He observed specimens in North America and found that the leaves were slightly wider toward the west, but the difference was not sufficient to distinguish a species (Russell Citation1956). In this study, the plastid genome of V. selkirkii was determined for the first time, and phylogenetic analysis was performed to identify the relationships within Viola.
DNA was extracted using fresh leaves from Mt. Il-san, Gangwon-do Province (N38° 11′ 17.0″, E127° 47′ 48.0″). The voucher specimen was deposited at the Kangwon National University Herbarium (KWNU, https://biology.kangwon.ac.kr/biology, Ki-Oug Yoo, [email protected]) under the voucher no. KWNU 98801. DNA extraction was performed using a DNA Plant Mini Kit (Qiagen Inc., Valencia, CA). The extracted DNA was sequenced using the Illumina MiSeq platform (Illumina Inc., San Diego, CA), and paired-end reads with an average read length of 301 bp were identified from the 3,364,589 raw reads. We used the Map to Reference function in Geneious 7.1.9 (Biomatters Ltd., Auckland, New Zealand) to exclude nuclear and mitochondrial reads based on the published plastid genome of V. seoulensis Nakai 1918 (GenBank accession no. KP749924). Then, de novo assembly was implemented using Geneious 7.1.9, and a total of 378,781 reads were aligned. The complete plastid sequence was annotated based on the online program GeSeq (Tillich et al. Citation2017) coupled with manual correction for the start and stop codons. Additionally, we referred to published Viola plastid genomes (GenBank accession nos. KP749924, MH229816, MH229819, and MT012304).
The complete plastid genome of V. selkirkii (GenBank accession no. MW448361) has a total length of 156,774 bp (GC content: 36.3%). This genome has a typical quadripartite structure with a large single-copy (LSC) region of 85,930 bp, a small single-copy (SSC) region of 17,982 bp, and two inverted repeats (IRs) of 26,431 bp each. The plastid genome of V. selkirkii has a total of 131 genes, with 84 protein-coding genes, 37 tRNA genes, eight rRNA genes, and two pseudogenes. The IR regions contain 17 duplicated genes (ndhB, rpl2, rpl23, rps7, rps12, ycf2, ycf15, rrn16, rrn23, rrn4.5, rrn5, trnA-UGC, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC). Within the plastid genome of V. selkirkii, eight protein-coding genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, and rpoC1) and six tRNA genes (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) have one intron each, and three genes (clpP, rps12, and ycf3) include two introns.
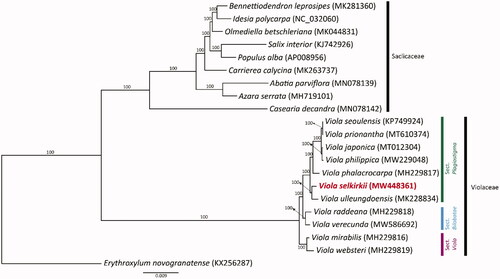
To determine the phylogenetic location of V. selkirkii in Viola, we conducted a phylogenetic analysis. A total of 77 protein-coding genes from 11 Violaceae species, nine Salicaceae species, and one outgroup (Erythroxylum novogranatense) were aligned by MAFFT (Katoh et al. Citation2002). Phylogenetic analyses were conducted with the maximum-likelihood (ML) method using RAxML v.7.4.2 with 1000 bootstrap replicates and the GTR + I model (Stamatakis Citation2006). The intraspecific classification system followed a study by Marcussen et al. (Citation2012). The phylogenetic analysis results () showed that Viola had a strongly supported monophyly (BS = 100) and formed a clade distinct from Salicaceae. V. selkirkii was closely related to V. ulleungdoensis and was placed in a basal position within sect. Plagiostigma. Our results contribute to the clear identification of the phylogenetic position of Viola and the phylogenetic relationships of other genera within Violaceae.
Author contributions
All authors were involved in the conception and design. Data collection: AG and KY; interpretation of data: AG and KC; draft manuscript preparation: AG. All authors reviewed the results and approved the final version, and all authors agree to be accountable for all aspects of the work.
Ethics statement
The plant material did not require any ethical or institutional approvals because this species is not on the list of endangered wildlife in Korea. Research on the plant (wild), including the collection of plant material, was carried out in accordance with guidelines provided by our institution, Kangwon National University, and national or international regulations.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that obtained at this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number MW448361. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA817383, SRR18352840, and SAMN26764828, respectively.
Additional information
Funding
References
- Brainerd E. 1921. Violets of North America. Vermont Agric Exp Stat Bull. 224:74.
- Cheon KS, Kim DK, Kim KA, Yoo KO. 2020. The complete chloroplast genome sequence of Viola japonica (Violaceae). Mitochondrial DNA Part B. 5(2):1297–1298.
- Clausen J. 1964. Cytotaxonomy and distributional ecology of Western North American violets. Madroño. 17(6):173–197.
- Katoh K, Misawa K, Kuma KI, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Kim KS. 1986. Studies of comparative morphology on the Korean Viola species [Ph.D. thesis]. Suwon: Sung Kyun Kwan University.
- Marcussen T, Jakobsen KS, Danihelka J, Ballard HE, Blaxland K, Brysting AK, Oxelman B. 2012. Inferring species networks from gene trees in high-polyploid North American and Hawaiian violets (Viola, Violaceae). Syst Biol. 61(1):107–126.
- Russell NH. 1956. Morphological variation in Viola selkirkii Pursh. Castanea. 21(2):53–62.
- Stamatakis A. 2006. RaxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wahlert GA, Marcussen T, de Paula-Souza J, Feng M, Ballard HE. 2014. A phylogeny of the Violaceae (Malpighiales) inferred from plastid DNA sequences: implications for generic diversity and intrafamilial classification. Syst Bot. 39(1):239–252.
- Yoo KO, Jang SK. 2010. Infrageneric relationships of Korean Viola based on eight chloroplast markers. J Syst Evol. 48(6):474–481.