Abstract
Thalictrum viscosum W.T.Wang & S.H.Wang, 1979 is a flowering plant species in family Ranunculaceae that is endemic to Yunnan province of China. To facilitate genetic study of T. viscosum, we de novo assembled and annotated the complete chloroplast (cp) genome of T. viscosum for the first time. The total length of the cp genome of T. viscosum was 155,984 bp, with a GC content of 38.4%. The T. viscosum cp genome had a typical quadripartite structure with a large single-copy region of 85,339 bp, a small single-copy region of 17,656 bp, and a pair of inverted repeat regions of 26,495 bp. The cp genome consisted of 133 genes, including 87 protein-coding genes, 38 transfer RNA genes, and eight ribosomal RNA genes. We performed phylogenetic analysis of T. viscosum with the maximum-likelihood phylogenetic tree and indicated that T. viscosum was closely related to T. cirrhosum and T. foeniculaceum.
Thalictrum viscosum W.T.Wang & S.H.Wang, 1979 is a flowering plant species in family Ranunculaceae. It grows in grassy places of the valleys or riversides and is endemic to Yunnan province of China (Zeng et al. Citation2020). Besides, the T. viscosum, as a medicinal herb with valuable botanical components that has received much attention (Chen et al. Citation2003; Khamidullina et al. Citation2006; Sharma et al. Citation2020). The chloroplast (cp) genome is a useful genetic tool for conservation research of endangered species (Arias et al. Citation2020; Song et al. Citation2022), but so far, there are no reports on the cp genome of T. viscosum. To facilitate further research on T. viscosum, we assembled and annotated the complete cp genome of T. viscosum for the first time. In addition, we performed phylogenetic analysis of T. viscosum to identify its phylogenetic relationships with other Ranunculaceae species.
The plant material of T. viscosum was collected in Kunming City, Yunnan Province, China (25°04′N, 102°42′E). We obtained approval for this research both from the local government and from the Kunming Institute of Botany, Chinese Academy of Sciences. A specimen was deposited at Qingdao University of Science and Technology (Chao Shi, [email protected]) under the voucher number TV202223. We washed the plant material with distilled water. Then, we extracted cpDNA from roughly 20 g of fresh plant leaves of T. viscosum using the more effective and improved high salt method (Shi et al. Citation2012). Subsequently, DNA quality was determined by spectrophotometric assessment. The high-quality DNA of T. viscosum was used to generate a genomic library for sequencing in Novogene Company (Beijing, China) with Illumina Hiseq 4000 platform. Low-quality reads and adaptors were clipped with Trimmomatic v0.36 (Bolger et al. Citation2014) and quality control was assessed by FastQC (Brown et al. Citation2017). Eventually, about 2.7 Gb of high-quality reads were generated. The complete cp genome of T. viscosum was assembled with NOVOPlasty v4.3.1 software (Dierckxsens et al. Citation2016). The assembly results were aligned by BLASTn (Johnson et al. Citation2008) to find the reference genome with the highest similarity and finally T. foeniculaceum (accession number NC_053570) was chosen as the reference sequence. The GeSeq software (Tillich et al. Citation2017) was used to annotate the protein-coding genes (PCGs), ribosomal RNA (rRNA) genes, and transfer RNA (tRNA) genes contained in the genome. Lastly, the annotation results were manually corrected against the reference genome by Sequin (Lehwark and Greiner Citation2019). The assembled T. viscosum cp genome sequence was submitted to the GenBank database under accession number MZ442609.
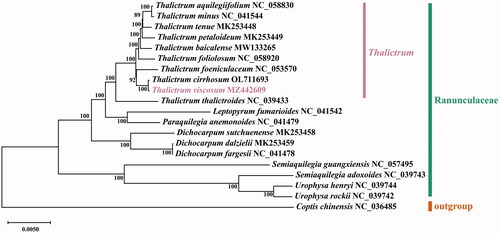
The total length of the T. viscosum cp genome was 155,984 bp, with a GC content of 38.4%. The makeup of the nucleotides was asymmetric (30.5% A, 19.6% C, 18.8% G, and 31.1% T). The T. viscosum cp genome had a typical quadripartite structure. A large single-copy (LSC) region of 85,339 bp and a small single-copy (SSC) region of 17,656 bp were separated by two equal 26,495 bp inverted repeat (IR) regions, IRa and IRb. The cp genome consisted of 133 genes, including 87 PCGs, 38 tRNA genes, and eight rRNA genes. A total of 23 genes with introns were annotated in the T. viscosum cp genome, with two genes (clpP1 and ycf3) having two introns and 21 genes having one intron. The trnK-UUU gene contained the largest intron, with a length of 2,521 bp, while the trnL-UAA gene contained the smallest intron, with a length of 531 bp. To determine the phylogenetic position of T. viscosum, we constructed a phylogenetic tree using the complete cp genomes of 20 Ranunculaceae species that have been released in the NCBI database and by using Coptis chinensis as an outgroup (). The complete cp genome sequences of 20 Ranunculaceae species were aligned using MAFFT v725 (Katoh and Standley Citation2013). The Modeltest (Posada and Crandall Citation1998) was used to determine the best nucleotide substitution model. The best model, GTR-GAMMA (GTR + G), was selected to construct the phylogenetic tree based on the Bayesian information criterion (BIC). The maximum-likelihood (ML) tree was constructed using MEGA-X software (Kumar et al. Citation2018). The ML phylogenetic tree indicated that T. viscosum was closely related to T. cirrhosum and T. foeniculaceum and support the conclusion of the earlier study (He et al. Citation2021) that T. baicalense was closely related to T. tenue, T. minus, and T. petaloideum.
Author contributions
Haohong Cai: conceptualization, writing – original draft. Wenbo Shi: formal analysis, software. Weicai Song: methodology, data curation. Weiqi Han: data curation, formal analysis. Shuo Wang: data curation, resources, writing – reviewing and editing. All authors have read and approved the final manuscript.
Acknowledgements
We would like to acknowledge the NGS service provided by Beijing-based Novogene, which has been instrumental in the implementation of the project. We are grateful to Chao Shi and Zimeng Chen for their guidance and assistance in the assembly of the chloroplast genome of Thalictrum viscosum in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession MZ442609. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA818965, SRR18441631, and SAMN26892146, respectively.
Additional information
Funding
References
- Arias T, Riaño-Pachón DM, Di Stilio V. 2020. Facilitating candidate gene discovery in an emerging model plant lineage: transcriptomic and genomic resources for Thalictrum (Ranunculaceae). Appl Plant Sci. 9(1):e11407.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Brown J, Pirrung M, McCue LA. 2017. FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics. 33(19):3137–3139.
- Chen SB, Chen SL, Xiao PG. 2003. Ethnopharmacological investigations on Thalictrum plants in China. J Asian Nat Prod Res. 5(4):263–271.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- He YF, Wang RN, Gai XY, Lin PC, Li YX, Wang JL. 2021. The complete chloroplast genome of Thalictrum baicalense Turcz. ex Ledeb. Mitochondrial DNA Part B. 6(2):437–438.
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res. 36(Web Server):W5–W9.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Khamidullina EA, Gromova AS, Lutsky VI, Owen NL. 2006. Natural products from medicinal plants: non-alkaloidal natural constituents of the Thalictrum species. Nat Prod Rep. 23(1):117–129.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lehwark P, Greiner S. 2019. GB2sequin – a file converter preparing custom GenBank files for database submission. Genomics. 111(4):759–761.
- Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics. 14(9):817–818.
- Sharma N, Kumar V, Chopra MP, Sourirajan A, Dev K, El-Shazly M. 2020. Thalictrum foliolosum: a lesser unexplored medicinal herb from the Himalayan region as a source of valuable benzyl isoquinoline alkaloids. J Ethnopharmacol. 255:112736.
- Shi C, Hu N, Huang H, Gao J, Zhao YJ, Gao LZ. 2012. An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. PLOS One. 7(2):e31468.
- Song WC, Ji CX, Chen ZM, Cai HH, Wu XM, Shi C, Wang S. 2022. Comparative analysis the complete chloroplast genomes of nine Musa species: genomic features, comparative analysis, and phylogenetic implications. Front Plant Sci. 13:832884.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Zeng YP, Yuan Q, Yang QE. 2020. Thalictrum panzhihuaense (Ranunculaceae) is placed in synonymy with T. glandulosissimum, with reduction of T. tsaii to T. glandulosissimum var. tsaii. Phytotaxa. 452(2):137–154.