Abstract
Ficus sarmentosa is a common climbing fig tree in East Asia in Moraceae, and its particular geographical distributed pattern and on-going radiation evolution make it significant to explore evolutionary history and biogeography of Ficus. In this work, the first complete chloroplast genome of F. sarmentosa was reported using Illumina NovaSeq high-throughput sequencing data. Totally, the whole chloroplast genome of F. sarmentosa is 160,183 bp in length, which includes large single-copy region of 88,307 bp, small single-copy region of 20,080 bp, and two pairs of inverted repeat regions of 25,898 bp. In addition, GC content and microsatellite makers of the genome were explored. Lastly, phylogenetic analysis demonstrated the closest relationship between F. sarmentosa and F. pumila.
Ficus Linnaeus is the largest genus in Moraceae with extreme species diversity as well as ecologically and evolutionarily important on account of unique obligate mutualism to fig wasp, providing an ideal case for studies of speciation and co-evolution (Wang et al. Citation2021). Nevertheless, Ficus species that have available complete chloroplast genomes in open data repository such as GenBank are less than five percent, rendering that molecular data in this species-rich genus are now far insufficient to support the further researches. Ficus sarmentosa Buch.-Ham. ex J.E.Sm. 1810 is a widely distributed climbing fig tree throughout East Asia but unexpectedly absent in adjacent Southeast Asia region (Zhou and Gilbert Citation2003), in view of the latter is thought to be the distributed center and possible ancestral area of Ficus (Cruaud et al. Citation2012; Pederneiras et al. Citation2018). Additionally, F. sarmentosa possesses up to eight indistinguishable varieties and has numerous allies in subsect. Plagiostigma, which forms a species complex under rapid radiation evolution (Zhang et al. Citation2020). Therefore, studies of its genetic characteristics, including the chloroplast genome and phylogenetic analysis, are of the essence.
In the study, the sample of F. sarmentosa was obtained from Putuo Mountain in Zhoushan District, Zhejiang, China (122.40 E, 30.02 N) and the voucher specimen (collector: Lu Zou and Hong-Qing Li, collected number: ZZ063) was deposited in the herbarium of East China Normal University (HSNU, Curator: Rui-Liang Zhu, email: [email protected]). Total genomic DNA was extracted from dried leaves using CTAB method (Doyle and Doyle Citation1987). The shotgun library was prepared based on the total genomic DNA after shearing, end repair and adapter ligation. 2G clean paired-end data (deposited in SRA with the accession number SRR16894930) were obtained with Illumina NovaSeq PE150 platform (San Diego, CA). The chloroplast genome was assembled using GetOrangelle v1.7.5 (Jin et al. Citation2020) and annotated via PGA v1.0 (Qu et al. Citation2019) under default parameters. The complete chloroplast genome sequence was deposited in GenBank (accession number: OL415083).
Figure 1. The maximum-likelihood phylogenetic tree based on complete chloroplast genomes. Numbers above the branches indicated the values from 10,000 ultrafast bootstrap.
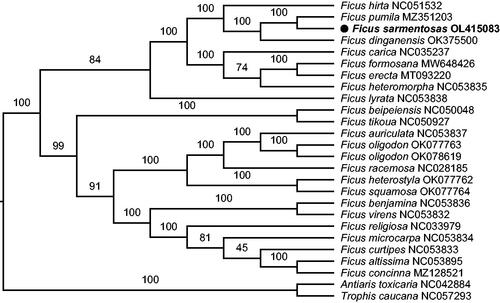
As a result, the complete genome sequence of F. sarmentosa is 160,183 bp in length, consisting of a large single-copy (LSC) of 88,307 bp, a small single-copy (SSC) of 20,080 bp, and two inverted repeats (IRs) of 25,898 bp. In addition, the overall GC content is 35.99%. Generally, 131 genes were annotated, which included 86 coding genes, 37 tRNA genes, and eight ribosomal RNA genes. What is more, 46 microsatellite loci with mononucleotide repeat unit, eight with dinucleotide unit and one with trinucleotide unit, in total 55 microsatellite loci were detected throughout the chloroplast genome via MISA-web (https://webblast.ipk-gatersleben.de/misa/) under default parameters, and these genetic resources are meaningful to the exploration of population genetics and evolutionary history in F. sarmentosa species complex. To investigate the phylogenetic placement of F. sarmentosa, the maximum-likelihood (ML) tree was reconstructed with 23 available chloroplast genomes of Ficus in GenBank via IQ-tree v2.1.3 (Nguyen et al. Citation2015) under 10,000 ultrafast bootstrap (Hoang et al. Citation2018), after aligned by MAFFT v7.490 (Katoh and Standley Citation2013). The related species Antiaris toxicaria (Pers.) Lesch. 1810 and Trophis caucana (Pittier) C.C.Berg 1988 were selected as outgroup. The ML tree showed that F. sarmentosa is close to F. pumila Linnaeus 1753 (), which is a species of the same subsection, consistent with pervious results (Zhang et al. Citation2020). The complete chloroplast genome of the widely distributed species F. sarmentosa will be promising for a better understanding of population structure, evolutionary history, and biogeography in Ficus.
Ethical approval
For all the lab experiments and data analyses in the study, no ethical approval or other permissions is necessary to be provided. Meanwhile, field work complies with local legislation, and no national or international law and regulations were violated.
Author contributions
De-Shun Zhang was involved in the conception and design of the study; Zhen Zhang performed experiments, data analysis, and preparation of manuscript draft; and both two authors took part in final approval of the paper and agreed to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. OL415083. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA771926, SRR16894930, and SAMN23012429, respectively.
Additional information
Funding
References
- Cruaud A, Rønsted N, Chantarasuwan B, Chou LS, Clement WL, Couloux A, Cousins B, Genson G, Harrison RD, Hanson PE, et al. 2012. An extreme case of plant-insect codiversification: figs and fig-pollinating wasps. Syst Biol. 61(6):1029–1047.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1–31.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Pederneiras LC, Gaglioti AL, Romaniuc-Neto S, Mansano VDF. 2018. The role of biogeographical barriers and bridges in determining divergent lineages in Ficus (Moraceae). Bot J Linn Soc. 187(4):594–613.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Wang G, Zhang X, Herre EA, McKey D, Machado CA, Yu WB, Cannon CH, Arnold ML, Pereira RA, Ming R, et al. 2021. Genomic evidence of prevalent hybridization throughout the evolutionary history of the fig-wasp pollination mutualism. Nat Commun. 12(1):1–14.
- Zhang Z, Wang XM, Liao S, Zhang JH, Li HQ. 2020. Phylogenetic reconstruction of Ficus subg. Synoecia and its allies (Moraceae), with implications on the origin of the climbing habit. Taxon. 69(5):927–945.
- Zhou ZK, Gilbert MG. 2003. Moraceae. In: Wu ZY, Raven PH, editors. Flora of China. Beijing; St. Louis: Science Press; Missouri Botanical Garden Press; p. 21–73.
