Abstract
We sequenced the complete mitochondrial genome of sand dollar Astriclypeus mannii (Verrill 1867) (Echinoidea: Astriclypeidae) occurring in the subtidal sand flat in Jeju Island off the south coast of Korea. The mitochondrial genome was 15,744 bp in length and contained 13 protein-coding genes (PCGs), 22 transfer RNAs, two ribosomal RNAs, and 140 nucleotides representing the putative control region. We reconstructed the concatenated phylogenetic tree based on 13 PCGs of 18 echinoderms, including A. mannii. From the maximum likelihood clustering, A. mannii was grouped in the order Echinolampadacea. The complete mitochondrial sequence of A. mannii for the first time in this study provide valuable insight in understanding the evolution and phylogenetic analysis of echinoids (sea urchins).
Common in subtidal sand flats, the sand dollar Astriclypeus mannii (=A. manni, Verrill 1867) has a wide distribution in the West Pacific Ocean ranging from Jeju Island off the south coast of Korea, Taiwan, and the Hainan Island in the South China Sea (Yulin Citation1998; Kang et al. Citation1999; Chao Citation2000). According to Kang and Choi (Kang and Choi Citation2002), A. mannii is a deposit feeder feeding on foraminifera, harpacticoid copepods, and diatoms in the sediment. Currently, five genera and eight species are in the family Astriclypeidae including A. mannii. However, mitochondrial genomes of the sand dollars in the family Astriclypeidae are yet to be available. Accordingly, we first analyzed the complete mitochondrial genome of A. mannii.
We collected A. mannii from subtidal sand flat in northern Jeju Island, Korea (33°29′07.40″N, 126°54′43.40″E). This area was not designated as a protected area by the government and no permit was required for the study. The type specimens with the voucher were deposited at the Library of Marine Samples of Korea Institute of Ocean Science and Technology (http://lims.kiost.ac.kr, Seung Won Jung, [email protected]) under the voucher number (B_S_MA_00031798). The total genomic DNA was extracted from the gonad tissue using a DNA isolation kit (Qiagen, Hilden, Germany) following the manufacturer's protocol. DNA libraries (insert size: 550 bp) were constructed by TruSeq DNA Nano TM kit (Illumina, Sandiego, CA). Genome sequencing was carried out using the Novaseq 6000 platform at DNAlink Inc., Korea, acquiring 121 million 151 bp paired-end raw reads (36.8 Gbp). Raw sequence data were trimmed with Trim Galore (ver. 0.6.6) (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore) to remove low-quality reads and adapter sequences. Mitogenome assembly was performed using mitoAssemble with k-mer size 31 implemented in MitoZ (ver. 2.3) (Meng et al. Citation2019). Genes were initially annotated with MITOS (Bernt et al. Citation2013) and MITOS2 (Donath et al. Citation2019) and manually modified with related species (Perseke et al. Citation2010).
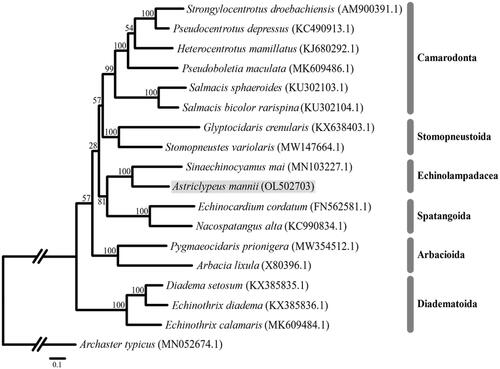
The complete mitogenome length of A. mannii is 15,744 bp (NCBI accession number: OL502703), similar to other published echinoids (Perseke et al. Citation2010). The mitogenome contains 37 genes, a conserved set of 13 protein-coding genes (PCGs), two ribosomal RNA genes (12S and 16S), 22 transfer RNA genes, and 140 nucleotides represent the putative control region. The overall base composition of the mitogenome is estimated to be A 31.38%, T 29.60%, C 23.54%, and G 15.49% with [A + T] content of 60.98%, which is most similar to Stomopneustes variolaris (59.77%) (Zhong et al. Citation2021). We reconstructed the concatenated phylogenetic tree using the maximum likelihood method with the GTR + F+ I + G4 substitution model with 1000 bootstrap replicates based on the 13 PCGs obtained from A. mannii and 16 echinoid species (all available sequences in order Diadematoida, Arbacioida, Spatangoida, Stomopneustoida, Echinolampadacea, and 6 representative species in Camarodonta) (). In the phylogenetic tree, A. mannii was grouped in order Echinolampadaceae and showed the same results as the current echinoid phylogenetic study (Mongiardino Koch et al. Citation2018; Lin et al. Citation2020). The complete mitochondrial sequence of A. mannii analyzed for the first time in this study could provide valuable database for the establishment of a phylogenetic analysis of echinoids (sea urchins).
Figure 1. Maximum likelihood (ML) phylogeny of 17 echinoid species (six Camarodonta, two Stomopneustoida, two Spatagoida, two Echinolampadacea including A. mannii, two Arbacioda, and three Diadematoida) based on the concatenated nucleotide sequences of 13 PCGs. Asteroidea (A. typicus) was used as the outgroup. Numbers on the branches indicate ML bootstrap percentages (1000 replicates). The Genbank accession numbers for the published sequences are incorporated. The gray box indicates the A. mannii analyzed in this study.
Author contributions
J.-S. Shin: Writing the original manuscript, Resources, Conceptualization. C.-u. Song: Data analysis, Methodology. H. Choi: Data analysis, Visualization. D.-H. Kang: Resources. K.K. Kwon: Project administration. S. Eyun: Writing revision and editing, Supervision of the genome analysis. K.-S. Choi: Writing revision and editing, Supervision of the project.
Disclosure statement
No potential competing interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. OL502703. The associated Bio-Project, SRA, and Bio-Sample numbers are PRJNA801414, SRR17794416, and SAMN25343171 respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogen. Evol. 69(2):313–319.
- Chao S-M. 2000. The irregular sea urchins (Echinodermata: Echinoidea) from Taiwan, with descriptions of six new records. Zool Stud. 39:250–265.
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552.
- Kang D-H, Choi K-S. 2002. Ecological studies on the sand dollar, Astriclypeus manni (Verril 1867) – feeding, density and locomotion. Korean J Environ Biol. 20:180–188.
- Kang D-H, Choi K-S, Chung S-C. 1999. An ecological study on the sand dollar, Astriclypeus manni (VERRIL 1867), in Hamdock, Cheju Korea. Korean J Fish Aquat Sci. 32:345–352.
- Lin JP, Tsai MH, Kroh A, Trautman A, Machado DJ, Chang LY, Reid R, Lin KT, Bronstein O, Lee SJ, et al. 2020. The first complete mitochondrial genome of the sand dollar Sinaechinocyamus mai (Echinoidea: Clypeasteroida). Genomics. 112(2):1686–1693.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Mongiardino Koch N, Coppard SE, Lessios HA, Briggs DE, Mooi R, Rouse GW. 2018. A phylogenomic resolution of the sea urchin tree of life. BMC Evol Biol. 18(1):1–18.
- Perseke M, Bernhard D, Fritzsch G, Brümmer F, Stadler PF, Schlegel M. 2010. Mitochondrial genome evolution in Ophiuroidea, Echinoidea, and Holothuroidea: insights in phylogenetic relationships of Echinodermata. Mol Phylogenet Evol. 56(1):201–211.
- Yulin L. 1998. The echinoderm fauna of Hainan Island. In: Brian M, editor. Proceedings of the 3rd international conference on the marine biology of the South China Sea. Hong Kong: Hong Kong University Press; p. 75–82.
- Zhong S, Zhao L, Huang L, Liu Y, Huang G. 2021. The first complete mitochondrial genome of Stomopneustes variolaris (Lamarck, 1816) from the Stomechinidae (Echinoidea: Stomopneustoida). Mitochondrial DNA B Resour. 6(8):2258–2259.
