Abstract
In this study, the complete chloroplast genome of Acer paihengii, a tree species native to China, was sequenced and assembled through second-generation sequencing. The complete chloroplast genome of A. paihengii is 155,967 bp in length with a typical quadripartite structure, encompassing 130 genes including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Phylogenetic analysis of 22 related species indicated that A. paihengii was more closely related to Acer coriaceifolium and Acer sino-oblongum.
Acer paihengii is a unique deciduous tree species with high ornamental value in China. It is mainly distributed in Yunnan Province, China, where it is a provincial key protected wild plant species (Zhou Citation2010; Qin et al. Citation2017). Most studies on Aceraceae plants in China mainly focus on genetic breeding, introduction and domestication, cultivation techniques, economic uses, and landscape ecological applications, among other aspects. However, few studies have focused on A. paihengii. The research involved in this species mainly focused on resource investigation and biodiversity.
The chloroplast (cp) genome is highly conserved among plants due to its semi-autonomous and maternal inheritance characteristics, and can thus provide important molecular data onto the characterization of plant systematic evolution and biogeography research (Gao et al. Citation2020; Yang et al. Citation2020). Here, the complete chloroplast genome of A. paihengii was assembled, annotated, and phylogenetically analyzed, thus providing crucial insights into the evolutionary relationship between A. paihengii and other members of the Acer genus such as Acer miaotaiense (Zhang et al. Citation2016), Acer buergerianum (Xu et al. Citation2017), Acer saccharum (Deng et al. Citation2019), Acer truncatum (Chen et al. Citation2019), and Acer tataricum subsp. ginnala (Yang et al. Citation2020).
Acer paihengii Fang was first mentioned in Act. Phytotax. Sin. 11: 169. 1966. Leaf samples of A. paihengii were collected from Henan Agricultural University (Henan, China, 113°67′E, 34°79′N) and the specimens were deposited in the Herbarium of Henan Agricultural University (http://bbg.henau.edu.cn/, Liu Yiping and E-mail: [email protected]) under the voucher number YJ20210325. Total genomic DNA was extracted using the OMEGA kit, after which an Illumina opposite-end library was constructed by Shanghai Yuanshen Biomedical Technology Co., Ltd. (Origingene, Shanghai, China) on an Illumina HiSeq TM sequencer (Illumina, San Diego, CA, USA). The raw data was approximately 5.26 G and low-quality sequences were filtered out to obtain clean and high-quality data. The chloroplast genome was assembled using the NOVOPlasty4.2 software (Nicolas et al. Citation2017). Gene annotation was performed using the PGA annotation software (https://github.com/quxiaojian/PGA) (Qu et al. Citation2019).
The whole length of the A. paihengii chloroplast genome (GenBank accession: MZ934750) is 155,967 bp, including a pair of 26,063 bp inverted repeat regions (IRa and IRb), a large single-copy (LSC) 85,798 bp region, and a small single-copy (SSC) 18,043 bp region. Furthermore, the total GC content of this circular DNA molecule was 35.62%. A total of 130 functional genes were annotated, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. These three types of genes accounted for 65.39, 28.46, and 6.15% of all annotated functional genes, respectively. Among them, a total of 15 genes (trnK-UUU, trnG, trnL-UAA, trnV-UAC, trnl-GUA, trnA-UGC, rps16, ropC1, atpF, petB, petD, rpl16, rpl2, ndhA, and ndhB) contained one intron. In contrast, clpP and rps12 possess two introns. Among which trnI-GUA, trnA-UGC, rpl2, ndhB, and rps12 exist as double copies.
To study the phylogenetic position of A. paihengii, 22 complete chloroplast genome sequences were downloaded from the NCBI GenBank. Sequence alignment was performed using MAFFT v7.158b (Katoh and Standley Citation2013). A phylogenetic tree was then generated via maximum likelihood analysis in RaxML (Stamatakis Citation2014). The development analysis results indicated that most nodes in the phylogenetic tree were strongly supported and all 23 Acer plants were clustered in an evolutionary branch. A. paihengii, A. coriaceifolium, and A. sino-oblongum clustered together, indicating a close evolutionary relationship (). In summary, the complete chloroplast genome of A. paihengii obtained in this study provides a robust basis for future phylogenetic studies of the Acer genus.
Figure 1. Phylogenetic tree of 23 complete chloroplast genome sequences of members of the order Sapindales. The numbers next to the nodes represent the bootstrap support values.
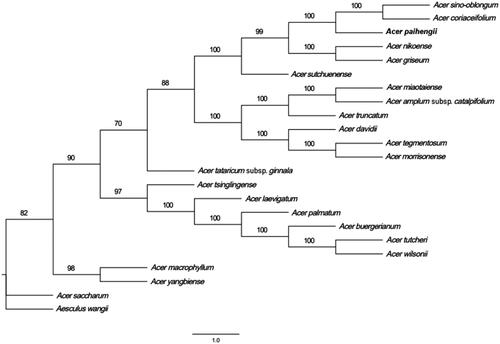
The following sequences were used: Acer truncatum NC_037211.1(Chen et al. Citation2019), Acer miaotaiense NC_030343.1 (Zhang et al. Citation2016), Acer amplum subsp. catalpifolium NC_041080.1 (Wang et al. Citation2019), Acer tataricum subsp. ginnala MN790641.1 (Yang et al. Citation2020), Acer wilsonii NC_040988.1, Acer tutcheri NC_051542.1 (Shi et al. Citation2020), Acer buergerianum NC_034744.1 (Xu et al. Citation2017), Acer palmatum NC_034932.1, Acer laevigatum NC_042443.1, Acer tsinglingense MN393475.1 (Dong et al. Citation2019), Acer yangbiense MN652924.1 (Ling and Zhang Citation2020), Acer macrophyllum NC_056217.1, Acer saccharum NC_051960.1 (Deng et al. Citation2019), Acer sutchuenense NC_049166.1 (Zhang et al. Citation2020), Acer paihengii MZ934750.1, Acer sino-oblongum NC_040106.1, Acer cinnamomifolium MN414240.1 (Chen et al. Citation2019), Acer nikoense NC_049165.1 (Fu et al. Citation2020), Acer griseum NC_034346.1 (Wang et al. Citation2017), Acer davidii NC_030331.1 (Jia et al. Citation2016), Acer morrisonense NC_029371.1 (Li et al. Citation2017), Acer tegmentosum MK942342.1 (Kim et al. Citation2019), Aesculus wangii NC_035955.1 (Zheng et al. Citation2018).
Ethical approval
We ensure that all experiments comply with ethical requirements. The research permission is provided by Henan Agricultural University.
Author contributions
Yiping Liu: Conceptualization; Project administration; Resources; Supervision; Review and editing. Yunru Zhai: Resources; Data curation; Software; Formal analysis; Original draft; Review and editing; Visualization. Dan He: Methodology; Conceptualization. Hongli Liu: Conceptualization; Review and editing. Man Zhang: Methodology; Conceptualization. Dezheng Kong: Supervision; Project administration; Funding acquisition.
Disclosure statement
No potential conflict of interest was reported by the author(s). We guarantee that this article has not been submitted to other journals at the same time, and that all content has not been published. All authors agree to the publication of the article and have no conflict of interest, financial or otherwise.
Data availability statement
The genome sequence data supporting the findings of this study are openly available in the NCBI GenBank database at https://www.ncbi.nlm.nih.gov/ under the accession no. MZ934750. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA767933, SRR16970252, and SAMN22253490, respectively.
Additional information
Funding
References
- Chen SJ, Liu BB, Zhang SX, Huang J. 2019. The complete chloroplast genome of Acer truncatum Bunge (Aceraceae). Mitochondrial DNA Part B. 4(1):607–608.
- Chen MH, Zhang HJ, Jiang M. 2019. The complete chloroplast genome sequence of Acer cinnamomifolium (Aceraceae), a plant species endemic to China. Mitochondrial DNA B Resour. 4(2):3450–3451.
- Deng X, Jiang ZX, Huang JC, Zhang XZ. 2019. Characterization of the complete chloroplast genome of sugar maple (Acer saccharum). Mitochondrial DNA B Resour. 5(1):21–22.
- Dong PB, Liu Y, Gao QY, Yang T, Chen XY, Yang JY, Shang QH, Fang MF. 2019. Characterization of the complete plastid genome of Acer tsinglingense, an endemic tree species in China. Mitochondrial DNA B Resour. 4(2):4065–4066.
- Fu QD, Yu XD, Xia XH, Zheng YQ, Zhang CH. 2020. Complete chloroplast genome sequence of Acer nikoense (Sapindaceae). Mitochondrial DNA Part B. 5(3):3136–3137.
- Gao J, Yu T, Li JQ. 2020. Phylogenetic and biogeographic study of Acer L. section Palmata Pax (Sapindaceae) based on three chloroplast DNA fragment sequences. Acta Ecologica Sinica. 40(17):5992–6000.
- Jia Y, Yang J, He YL, He Y, Niu C, Gong LL, Li ZH. 2016. Characterization of the whole chloroplast genome sequence of Acer davidii Franch (Aceraceae). Conservation Genet Resour. 8(2):141–143.
- Katoh K, Standley DM. 2013. Mafft multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim SC, Shin S, Lee MW, Lee JW. 2019. Complete chloroplast genome of Acer tegmentosum and phylogenetic analysis. Mitochondrial DNA B Resour. 4(2):2555–2556.
- Li ZH, Xie YS, Tao Z, Jia Y, He YL, Yang J. 2017. The complete chloroplast genome sequence of Acer morrisonense (Aceraceae). Mitochondrial DNA. Part A. Mitochondrial DNA A DNA Mapp Seq Anal. 28(3):309–310.
- Ling LZ, Zhang SD. 2020. The complete chloroplast genome of an endangered and endemic species, Acer yangbiense (Aceraceae). Mitochondrial DNA Part B. 5(1):224–225.
- Nicolas D, Patrick M, Guillaume S. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Qin HN, Yang Y, Dong SY, He Q, Jia Y, Zhao LN, Yu SX, Liu HY, Liu B, Yan YH, 1State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, et al. 2017. Threatened species list of China’s higher plants. Biodiv Sci. 25(7):696–744.
- Qu XJ, Michael JM, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Shi ZW, Sun B, Pei NC, Shi X. 2020. The complete chloroplast genome of Acer tutcheri Duthie (Acereae, Sapindaceae): an ornamental tree endemic to China. Mitochondrial DNA B Resour. 5(3):2686–2687.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang WC, Chen SY, Zhang XZ. 2017. The complete chloroplast genome of the Endangered Chinese paperbark maple, Acer griseum (Sapindaceae). Conservation Genet Resour. 9(4):527–529.
- Wang A, He J, Xu SJ, Peng LL, Zhao LC. 2019. The complete chloroplast genome of Acer catalpifolium (Sapindaceae), a rare and Endangered plant in Southwest China. Mitochondrial DNA Part B. 4(1):2037–2038.
- Xu JH, Wu HB, Gao LZ. 2017. The complete chloroplast genome sequence of the threatened trident maple Acer buergerianum (Aceraceae). Mitochondrial DNA B Resour. 2(1):273–274.
- Yang HX, Zha X, Cao SL, Wang Y, Gao F, Zhou YJ. 2020. Complete chloroplast genome sequence of Acer ginnala, an important ornamental tree. Mitochondrial DNA B Resour. 5(1):609–610.
- Zhang Y, Li B, Chen H, Wang YC. 2016. Characterization of the complete chloroplast genome of Acer miaotaiense (Sapindales: Aceraceae), a rare and vulnerable tree species endemic to China. Conservation Genet Resour. 8(4):383–385.
- Zhang W, Xia XH, Chen DF, Li JY, He W, Ma WB, Wu BX, Zheng YQ, Zhang CH. 2020. Complete chloroplast genome sequence of Acer sutchuenense subsp. tienchuanenge (Sapindaceae). Mitochondrial DNA Part B. 5(3):2886–2887.
- Zheng W, Wang W, Harris AJ, Xu XD. 2018. The complete chloroplast genome of vulnerable Aesculus wangii (Sapindaceae), a narrowly endemic tree in Yunnan, China. Conservation Genet Resour. 10(3):335–338.
- Zhou B. 2010. Revising the Yunnan Key protected wild plants list. Plant Diversity. 32(3):221–226.
