Abstract
Tripterygium wilfordii is a perennial vine plant with medicinal value and belongs to the family of Celastraceae. In this study, we sequenced and analyzed the complete chloroplast genome of T. wilfordii. The chloroplast genome was 156,700 bp in length with a GC content of 37.47%. It contained two inverted repeat (IR) regions of 26,461 bp; each region was separated by large single-copy and small single-copy regions of 85,409 bp and 18,369 bp, respectively. In total, we annotated 134 unique genes, consisting of 89 protein-encoding genes, 8 rRNAs and 37 tRNAs. Phylogenetic analysis revealed that T. wilfordii was sister to T. regelii in a clade of Tripterygiumii species that was sister to a clade of Euonymus species.
Tripterygium wilfordii Hook.fil. (1862), a typical representative of the genus Tripterygium, is a perennial vine plant that is widely distributed in East Asia (Liu et al. Citation2010). The roots of T. wilfordii, named ‘Leigongteng’ in Chinese, is used in traditional Chinese medicine. Pharmacological research has confirmed it has significant anti-inflammatory, immunosuppressive, antitumor, and other pharmacological benefits (Kang et al. Citation2021). Currently, several Chinese patent drugs derived from T. wilfordii have been approved by the China Food and Drug Administration for clinical usage in immunosuppression following organ transplantation and to treat autoimmune and inflammatory related diseases (Liu et al. Citation2020). However, wild T. wilfordii populations have diminished due to unregulated harvesting; therefore, it is necessary to develop genomic resources for this species to facilitate research. In this study, we sequenced and analyzed the complete chloroplast genome of T. wilfordii. This will provide useful information on the phylogeny and evolution of genus Tripterygium and aid studies to conserve the invaluable natural T. wilfordii populations.
The total genomic DNA was extracted by a modified CTAB method (Doyle and Doyle Citation1987) from fresh leaves of T. wilfordii collected from the Botanical Garden of Jiangsu Health Vocational College (Nanjing, China; N 32°5′2.333″, E 118°37′6.820″). A voucher specimen was deposited at the herbarium of Jiangsu Health Vocational College under the voucher number, FF20210720ZY-12 (https://www.jssmu.edu.cn/, contact person: Mr. Hu Xu, and email: [email protected]). The entire genome sequencing was implemented by Bio&Data Biotechnologies Inc. (Guangzhou, China). Following DNA extraction, we fragmented 1 μg of purified DNA by ultrasound on Covaris E220 (Covaris, Brighton, UK) and used it to set up ∼300 bp short-insert libraries. These qualified libraries were sequenced with PE150 bp on an BGISEQ-500 sequencer (Hefei Bio&Data Biotechnologies Inc.) according to the manufacturer’s instructions detailed in the previous literature (Huang et al. Citation2017). In total, 35.82 Mb clean reads were obtained and assembled de novo using NOVOplasty 2.7.2 (Dierckxsens et al. Citation2017). Annotation was performed using CPGAVAS2 (Shi et al. Citation2019) and Basic Local Alignment Search Tool (BLAST) (Altschul et al. Citation1997) searches.
The chloroplast genome of T. wilfordii was 156,700 bp in length, containing two inverted repeat (IR) regions of 26,461 bp; each region was separated by large single-copy (LSC) and small single-copy (SSC) regions of 85,409 and 18,369 bp, respectively. A total of 134 functional genes were predicted, including 89 protein-coding genes, 37 transfer ribonucleic acid (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. While most genes were in the single copy regions, 18 genes including 7 protein-coding genes, 4 rRNA genes, and 6 tRNA genes were duplicated in the IR regions. 19 genes had two exons and four genes (clpP, ycf3, and two rps12) contained three exons. The total sequenced GC content was 37.47%, while the corresponding values in the LSC, SSC, and IR regions were 35.34, 31.98, and 42.82%, respectively.
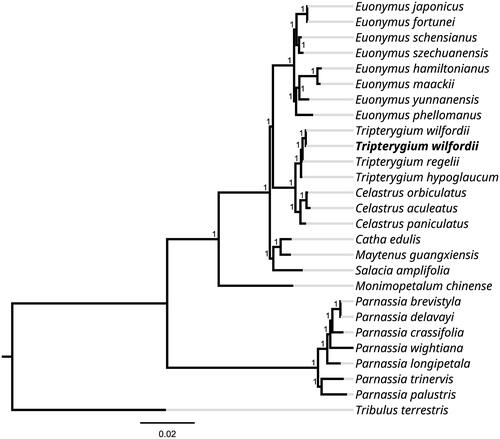
Alignment was carried out on the 27 chloroplast genome sequences using MAFFT version 7.0 to investigate phylogenetic relationships of T. wilfordii (Katoh and Standley Citation2013). A maximum likelihood (ML) tree was constructed using FastTree version 2.1.10 (Price et al. Citation2010). The results showed that among the Celastraceae species sampled, T. wilfordii was sister to T. regelii a clade containing all other sampled Tripterygium species. This clade was in turn sister to a clade containing the Euonymus species (). The newly disclosed chloroplast genome MN624264 of T. wilforgii was annotated using the same method as in this study. While the OK065822 and MN624264 plastomes have the same number of genes, the latter has a total length of 158,916 bp and 37.5% GC content owing to a 938 bp indel that was not detected in the T. wilfordii plastome we describe here. In conclusion, the complete chloroplast genome sequence of T. wilfordii will be useful for future research in conservation genetics and molecular-assisted breeding.
Figure 1. Phylogenetic tree inferred using the Maximum Likelihood (ML) method based on 27 representative species (with 1000 bootstrap repetitions). The following sequences were used: Tripterygium wilfordii OK065822 (in this study), Euonymus japonicus NC_028067 (Choi and Park Citation2016), Euonymus fortunei NC_057058 (unpublished), Euonymus schensianus NC_036019 (Wang et al. Citation2017), Euonymus szechuanensis NC_047463 (Wang et al. Citation2020), Euonymus hamiltonianus NC_037518 (unpublished), Euonymus maackii NC_057059 (unpublished), Euonymus yunnanensis MW770452 (unpublished), Euonymus phellomanus NC_057060 (unpublished), Tripterygium wilfordii MN624264 (unpublished), Tripterygium regelii MN624266 (unpublished), Tripterygium hypoglaucum MN624265 (unpublished), Celastrus orbiculatus MW316708 (unpublished), Celastrus aculeatus MW801026 (unpublished), Celastrus paniculatus OL804289 (unpublished), Catha edulis KT861471 (unpublished), Maytenus guangxiensis NC_047301 (Shi and Liu Citation2020), Salacia amplifolia NC_047214 (Lin et al. Citation2019), Monimopetalum chinense MK450440 (Pan et al. Citation2019), Parnassia brevistyla MG792145 (Xia et al. Citation2018), Parnassia delavayi MK580540 (unpublished), Parnassia crassifolia MK580538 (unpublished), Parnassia wightiana MN398191 (Li et al. Citation2019), Parnassia longipetala MK580539 (unpublished), Parnassia trinervis NC_043951 (unpublished), Parnassia palustris NC_045280 (Yu et al. Citation2019), Tribulus terrestris MN164624 (Yan et al. Citation2019).
Ethical approval
Research on plants (either cultivated or wild), including the collection of plant material, was undertaken in compliance with relevant institutional, national, and international guidelines and legislation.
CRediT authorship statement
Yuan Zhong: Writing – Original Draft, methodology, formal analysis. Jingzheng Zhang: Resources. Zhenzhen Bao: Conceptualization, methodology.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in the GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/nuccore/OK065822.1/), under the accession no. OK065822. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA761540, SRR15817515, and SAMN21354213, respectively.
Additional information
Funding
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402.
- Choi KS, Park S. 2016. The complete chloroplast genome sequence of Euonymus japonicus (Celastraceae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3577–3578.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang J, Liang X, Xuan Y, Geng C, Li Y, Lu H, Qu S, Mei X, Chen H, Yu T, et al. 2017. A reference human genome dataset of the BGISEQ-500 sequencer. Gigascience. 6(5):1–9.
- Kang BY, Zhao XT, Yang YL, Meng F, Li CX, Li XL, Chen TC. 2021. Pharmacological action and clinical application of Leigongteng (Tripterygium wilfordii). Chin Arch Tradit Chin Med. 39:102–106.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li J, Yang Q, Xu B, Liu ZL. 2019. The complete chloroplast genome of Parnassia wightiana (Celastraceae). Mitochondrial DNA B Resour. 4(2):3987–3988.
- Lin R, Zhao K, Wang H, Zhu Z, Wang H. 2019. Complete plastome sequence of Salacia amplifolia (Celastraceae): an endemic shrub in Hainan, China. Mitochondrial DNA B Resour. 4(1):1977–1978.
- Liu RP, Li XJY, Huang NN, Fan MY, Sun R. 2020. Toxicity of traditional Chinese medicine herbal and mineral products. Adv Pharmacol. 87:301–346.
- Liu WP, Liu SX, Tang HZ, Bai M, Wang LL, Liu Y. 2010. New research progress of Tripterygium wilfordii. Chin Tradit Herb Drugs. 41:1215–1218.
- Pan J, Fang Z, Ji DM, Gu TH, Cheng JS, Yu XM, Li S, Ye FR. 2019. Complete chloroplast genome of the wild-type Monimopetalum chinense (Celastraceae). Mitochondrial DNA B Resour. 4(2):2383–2384.
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLOS One. 5(3):e9490.
- Shi LC, Chen HM, Jiang M, Wang LQ, Wu X, Huang LF, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Shi YC, Liu BB. 2020. Complete chloroplast genome sequence of Maytenus guangxiensis (Celastraceae), a rare and Critically Endangered species endemic to China. Mitochondrial DNA B Resour. 5(1):536–537.
- Wang WC, Chen SY, Zhang XZ. 2017. Characterization of the complete chloroplast genome of the golden crane butterfly, Euonymus schensianus (Celastraceae). Conservation Genet Resour. 9(4):545–547.
- Wang X, Li H, Zheng M, Jiang J. 2020. The complete chloroplast genome of Euonymus szechuanensis. Mitochondrial DNA B Resour. 5(2):1130–1131.
- Xia M, Zhang F, Rao H, Chi X, Khan G, Zhang Y, Yu J, Chen S. 2018. Complete chloroplast genome sequence of Parnassia brevistyla (Celastraceae) and phylogenetic analysis with related species. Mitochondrial DNA B Resour. 3(2):1187–1188.
- Yan J, Zhang N, Duan Y. 2019. The complete chloroplast genome sequence of Tribulus terrestris, an important traditional Chinese medicine. Mitochondrial DNA B Resour. 4(2):3108–3109.
- Yu H, Guo F, Liu D, Zhang Z. 2019. Complete chloroplast genome sequence of Parnassia palustris (Celastraceae). Mitochondrial DNA B Resour. 4(1):1503–1504.
