Abstract
The complete mitochondrial genome of the clam Corbicula japonica is 17,432 bp in length. The sequence consists of 13 protein-coding, 2 ribosomal RNAs, and 22 transfer RNA genes (GenBank accession no. MZ895053). The proportion of base-pairs in C. japonica are A + T (70.5%) and G + C (29.5%). Phylogenetic analysis reveal C. japonica to be sister species to C. fluminea within the monophyletic genus Corbicula, with high support. This study is helpful to the classification of the brackish water clam C. japonica, which is difficult to identify during early development owing to variation of shell morphology.
Keywords:
The brackish water clam Corbicula japonica Prime, 1864, a filter-feeding bivalve, is widely distributed in rivers and brackish waters throughout the Asian region, and it is a commercially important species for food and inland fishery resource in Korea (Park et al. Citation2004; Koyama et al. Citation2015; Xie et al. Citation2018). Aquatic C. japonica clams inhabiting brackish waters are generally exposed to salinity changes and can accumulate water contaminants under highly stressful environments (Koyama et al. Citation2015; Zhang et al. Citation2020). The clams, such as C. japonica as well as Corbicula fluminea, have been proposed as a good bio-indicator for the assessment of aquatic environments, owing to their rapid growth and bioaccumulation (Koyama et al. Citation2015; Bertucci et al. Citation2018; Domingues et al. Citation2020). However, the classification of Asian Corbicula has been greatly complicated by the extraordinary morphological variation in shell characteristics (Park et al. Citation2004). Mitochondrial DNA (mtDNA) sequences such as those of the cytochrome c oxidase subunit I (COI) gene have been used to elucidate the phylogeny of Cyrenidae (Haponski and Foighil Citation2019). However, there are only limited number of completed mtDNA sequences in the Cyrenidae family (Wu et al. Citation2019; Zhang et al. Citation2019). In the present study, we determined the complete mitochondrial genome sequence of C. japonica collected from an estuary using Illumina next-generation sequencing. Specimens of C. japonica were collected from the estuarine area of Gwangyang Bay, Yeosu, South Korea (N 34°58.45′, E 127°46.02′) on September 2019. The article follows the ARRIVE guidelines (https://arrive-guidelines.org/). Genomic DNA was extracted from C. japonica whole body using the DNeasy blood & tissue kit (Qiagen, Valencia, CA, USA). The voucher specimen (CNUISI-021020130) was deposited at the Specimen Museum of Fisheries Science Institute, Chonnam National University (KY Park, [email protected]). DNA sequencing using Illumina HiSeq4000 was carried out in Macrogen Inc., (Seoul, Korea). De novo assembly of cleaned reads was performed by various k-mer using SPAdes v.3.13.0 (Bankevich et al. Citation2012). After assembly, MitoZ (v.2.3), a Python3-based toolkit, was used for annotation (Meng et al. Citation2019).
The mitochondrial genome sequence of C. japonica is available at the National Center for Biotechnology Information (NCBI) database (GenBank accession number MZ895053). The complete sequence of the mtDNA of C. japonica is 17,432 bp and comprises 37 genes, including 13 protein-coding genes (PCGs), 22 tRNAs, and 2 rRNAs. All genes located on the same positive strand, compared with the majority of bivalves. The A + T base content (70.5%) was higher than the G + C content (29.5%). Nucleotide base composition is as follows: A (27.5%), C (9%), G (20.5%), and T (43%). The gene order is same with C. fluminea. A total of 22 tRNA sequences were found throughout the mitogenome of C. japonica, which identified based on their respective anticodons and secondary structures, ranged in length from 62 (trnR, trnH, trnW) to 68 bp (trnM). In the two rRNAs, 12 s rRNA (rrnS) gene is 868 bp in length, which lies between the trnT and trnM genes, whereas 16 s rRNA (rrnL) gene is 1236 bp in length lying between the cob and atp8 genes. The start codon of ATP8, ND2, ND6 was ATG, that of COX1, COX2, CYTB, ND3 was ATT, that of ND4, ND4L was ATA, that of ATP6, ND1 was GTG, and that of ND5, COX3 was TTG. Two types of terminal codons (TAA and TAG) are used in all genes of PCGs.
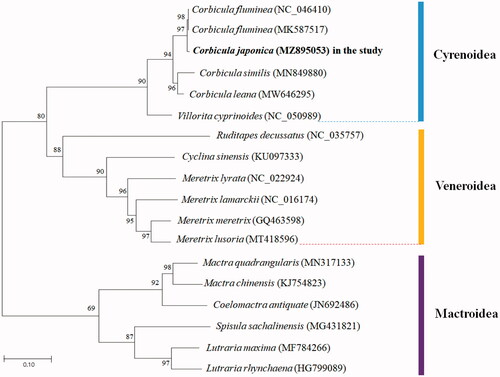
To analyze the phylogenetic relationships of C. japonica and other 17 mitochondrial genomes from Cyrenoidea, Veneroidea, and Mactroidea superfamilies in Venerida, phylogenetic trees were obtained using maximum-likelihood analysis based on all gene sequences by MEGA-X software (Kumar et al. Citation2018) (). The result of phylogenetic analysis showed that C. japonica formed a well-supported clade with C. fluminea, C. similis, and C. leana in Cyrenidae. C. japonica emerges as sister species to C. fluminea.
Ethical approval
Experiments were performed in accordance with the guidelines and regulations of the Animal Care and Use Committee of Chonnam National University (Yeosu, South Korea). This study did not involve Endangered or protected species.
Author contributions
KP and ISK contributed to the conception and design of the study, analysis, and interpretation of the data. KP wrote the first version of the manuscript. KP and ISK critically reviewed the article regarding its intellectual content. KP and ISK collected biological samples. All authors read, discussed, and approved the final version and all authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ895053. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA803933, SRR17952277 and SAMN25690507, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bertucci A, Pierron F, Gourves PY, Klopp C, Lagarde G, Pereto C, Dufour V, Gonzalez P, Coynel A, Budzinski H, et al. 2018. Whole-transcriptome response to wastewater treatment plant and stormwater effluents in the Asian clam, Corbicula fluminea. Ecotoxicol Environ Saf. 165:96–106.
- Domingues A, Rosa IC, da Costa JP, Rocha-Santos TA, Gonçalves FJ, Pereira R, Pereira JL. 2020. Potential of the bivalve Corbicula fluminea for the remediation of olive oil wastewaters. J Clean Prod. 252:119773.
- Haponski AE, Foighil DÓ. 2019. Phylogenomic analyses confirm a novel invasive North American Corbicula (Bivalvia: Cyrenidae) lineage. Peer J. 7:e7484.
- Koyama H, Okamoto S, Watanabe N, Hoshino N, Jimbo M, Yasumoto K, Watabe S. 2015. Dynamic changes in the accumulation of metabolites in brackish water clam Corbicula japonica associated with alternation of salinity. Comp Biochem Physiol B Biochem Mol Biol. 181:59–70.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: A toolkit for mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Park JK, Choe BL, Eom KS. 2004. Two mitochondrial lineages in Korean freshwater Corbicula (Corbiculidae: bivalvia). Mol Cells. 17(3):410–414.
- Wu CD, Zhang JQ, Bai ZY, Wang GL, Ge JY, Sun ML. 2019. The complete mitochondrial genome of the two kinds of Corbicula fluminea (black and yellow). Mitochondrial DNA B. 4(1):1383–1384.
- Xie Y, Chen H, Zheng S, Zhang X, Mu S. 2018. Molecular characterization of cu/Zn SOD gene in Asian clam Corbicula fluminea and mRNA expression and enzymatic activity modulation induced by metals. Gene. 663:189–195.
- Zhang T, Yan Z, Zheng X, Wang S, Fan J, Liu Z. 2020. Effects of acute ammonia toxicity on oxidative stress, DNA damage and apoptosis in digestive gland and gill of Asian clam (Corbicula fluminea). Fish Shellfish Immunol. 99:514–525.
- Zhang T, Yin J, Tang S, Li D, Liu X, Gu X, Liu Y. 2019. The complete mitogenome of clam Corbicula fluminea determined using next-generation and PacBio sequencing. Mitochondrial DNA B. 4(1):1660–1661.