Abstract
Ixodes ovatus is referred to as an obligatory blood-sucking ectoparasite that is capable to infest both humans and animals. In the present study, the complete mitochondrial genome of I. ovatus was sequenced and analyzed using next-generation sequencing (NGS) technology. With a size of 14,520 bp, the entire mitogenome contains 37 genes including 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), and 3 control regions (D-loops). Based on the 13 PCGs nucleotide sequences, the phylogenetic relationship of I. ovatus was analyzed using Maximum-likelihood. As suggested by the results of the obtained phylogenety, I. ovatus is most closely associated with Ixodes hexagonus. This study is expected to promote further studies on the evolution of Ixodidae.
Ixodes ovatus (Neumann, 1899) is a species of hard tick with a wide geographical distribution across China, Taiwan, Korea, Japan, Burma, and Thailand (Harry Hoogstraal Citation1973). With cows, horses, donkeys, deer, and sheep as known hosts, I. ovatus can accidentally parasitize humans, thus spreading a wide variety of severe animal and human pathogens, including Anaplasma phagocytophilum, Borrelia miyamotoi, Borrelia garinii, and Babesia spp., etc. (Shimada Citation2003). Therefore, correct identification of tick species is essential for disease control (Shimada Citation2003). However, conventional morphological identification requires extensive experience, which may restrict its applications. Since the mitochondrial genome is regarded as an effective genetic marker for species identification, sequencing of their complete mitochondrial genomes is beneficial to the identification and classification of ticks (Taanman Citation1999).
In this study, I. ovatus samples were collected in March 2021 from Tue Village, Nujiang City, Yunnan Province, China (98°48′59.84″E, 26°34′20.56″N) and then preserved in 75% ethanol. The recognition of samples was conducted by Professor Chunhong Du based on the morphological characteristics (Lu et al. Citation2021). Then, the specimen was deposited in Parasitological Museum, Dali University NO. DLUP2103 (URL: http://www.dali.edu.cn/jcyxy/xkpt/jcyxsyjxzx/6431.htm, Contact person: Xing Yang, [email protected]). The entire DNA was extracted using the standard CTAB technique, for storage at −20 °C before use (Lu et al. Citation2021). The mitochondrial genome was sequenced on the Illumina NovaSeq platform (Shanghai Personal Biotechnology Co, Ltd) which was assembled using A5-miseq software (Coil et al. Citation2015), and genome annotations were performed using the Swiss-Prot web server (http://www.ebi.ac.uk/uniprot/).
The mitochondrial genome of I. ovatus was determined as 14,520 bp (Genbank accession no. OM317739), involving 13 PCGs (cytb, nad1-6, atp8, nad4L, cox1-3, and atp6), 22 tRNAs, 2 rRNAs. The genetic order of the I. ovatus was identical to hard ticks. The entire base composition of the I. ovatus mitochondrial genome was determined as 37.54% A, 37.39% T, 16.24% C, 8.83% G. The size of I. ovatus small submit rRNA and large submit rRNA was 657 and 1,190 bp, respectively. It was discovered that the length of 22 tRNAs varied from 55 bp (tRNA-Ser) to 71 bp (tRNA-Lys), with 13 tRNAs encoded on the plus-strand (Thomas et al. Citation2013).
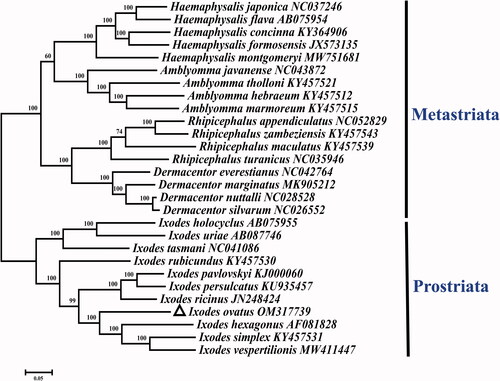
The GTR + G + I model was applied as the suitable model for sequencing, and the maximum-likelihood method was adopted through the MEGA 7.0 software with 1000 bootstrap replicates (Kumar et al. Citation2016). As shown in , the phylogenetic tree included complete mitogenomes sequences of 28 Ixodidae species previously published on GenBank. As revealed by the phylogenetic analysis, the obtained tree is divided into two phylogroups: Metastriata and Prostriata (Ciloglu et al. Citation2021). Additionally, it is shown that I. ovatus and all the other species within the genus Ixodes cluster in a branch with high statistical support, confirming I. ovatus within the genus Ixodes. The complete mitochondrial genome of I. ovatus provides an important molecular resource for further study on the phylogeny of the genus Ixodes and of Ixodidae (Kelava et al. Citation2021).
Figure 1. Maximum-likelihood (ML) phylogeny of 28 species of the family Ixodidae based on the 13 concatenated nucleotide sequences of protein-coding genes (PCGs), utilizing the GTR + G + I model and after 1,000 bootstrap replications. The black triangle sign represents the species in this study. Bootstrap support values are shown above the nodes.
Ethical approval
This study was approved by the Administration Committee of Experimental Animals, Dali University, Yunnan Province, China.
Author contributions
DDJ conceived the study and wrote the manuscript. XYL carried out the experiments and analyzed the data. CHD,SBH and ZPH contributed to the collection of I. ovatus and discussions, XY is responsible for the interpretation of experimental data, critical revision of important knowledge content and final approval of the version to be published.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov. The accession number of the complete mitochondrial genome is OM317739. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA820144, SRR18494365 and SAMN26982089, respectively.
Additional information
Funding
References
- Ciloglu A, Ibis O, Yildirim A, Aktas M, Duzlu O, Onder Z, Simsek E, Yetismis G, Ellis VA, Inci A. 2021. Complete mitochondrial genome characterization and phylogenetic analyses of the main vector of Crimean-Congo haemorrhagic fever virus: Hyalomma marginatum Koch, 1844. Ticks Tick Borne Dis. 12(5):101736.
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589.
- Hoogstraal H, Clifford CM, Saito Y, Keirans JE. 1973. Ixodes (partipalpiger) ovatus Neumann, subgen. J Med Entomol. 10(2):157–164.
- Kelava S, Mans BJ, Shao R, Moustafa M, Matsuno K, Takano A, Kawabata H, Sato K, Fujita H, Ze C, et al. 2021. Phylogenies from mitochondrial genomes of 120 species of ticks: Insights into the evolution of the families of ticks and of the genus Amblyomma. Ticks Tick Borne Dis. 12(1):101577.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lu X, Jiang D, Du C, Yang X. 2021. The complete mitochondrial genome of Haemaphysalis montgomeryi. Mitochondr DNA B Resour. 6(8):2233–2234.
- Lu X, Zuo X, Jiang D, Yang X. 2021. The complete mitochondrial genome of Ixodes vespertilionis (Acari: Ixodidae). Mitochondr DNA B Resour. 6(10):3001–3003.
- Shimada Y, Inokuma H, Beppu T, Okuda M, Onishi T. 2003. Survey 135 of Ixodes tick species on domestic. Vet Parasitol. 111(2-3):231–239.
- Shimada Y, Inokuma H, Beppu T, Okuda M, Onishi T. 2003. Survey of Ixodes tick species on domestic. Vet Parasitol. 111(2-3):231–239.
- Taanman J. 1999. The mitochondrial genome: structure, transcription, translation and replication. Netherlands: Elsevier B.V.; p. 103–123.
- Thomas D, Burger A, Renfu Shao B, Stephen C, Barker A. 2013. Phylogenetic analysis of the mitochondrial genomes and nuclear rRNA genes of ticks reveals a deep phylogenetic structure within the genus Haemaphysalis and further elucidates the polyphyly of the genus Amblyomma with respect to Amblyomma sphenodonti and Amblyomma elaphense. Ticks Tick Borne Dis. 4(4):265–274.
